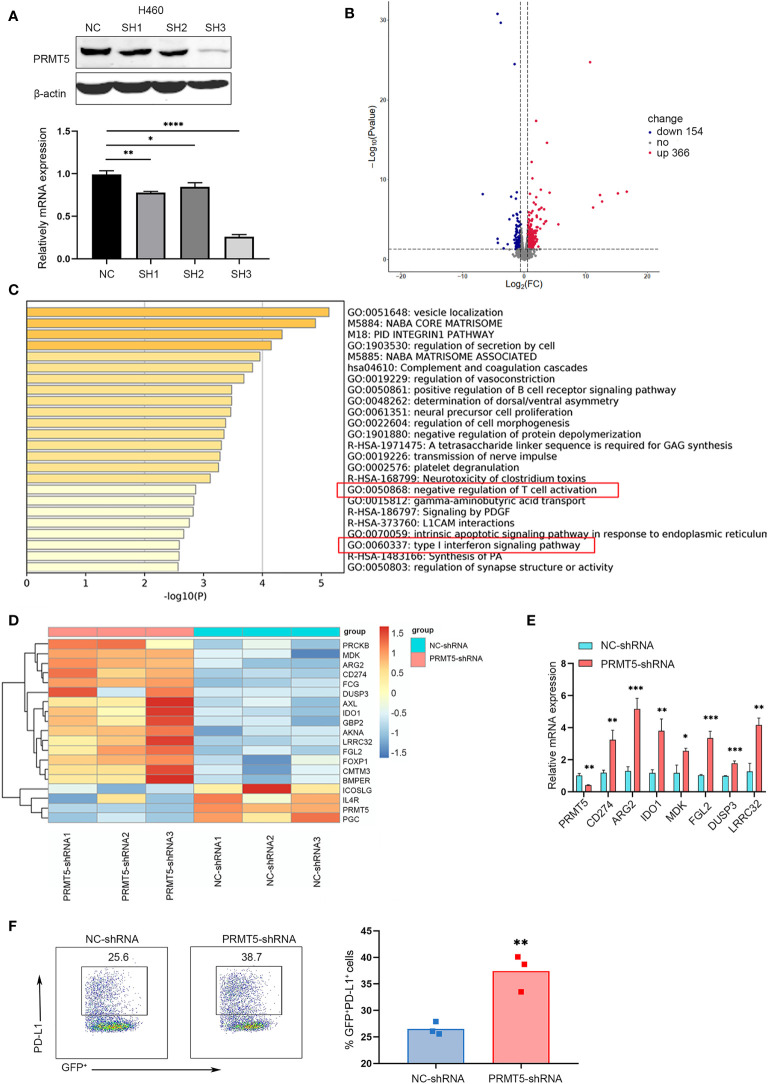
Figure 1.
Specific knockdown of PRMT5 in the human lung cancer cell line is related to negative regulation of T cell activation. (A) Three shRNA sequences targeting PRMT5 were designed. PRMT5-specific or control shRNA was transfected into the NCI-H460 human lung cancer line, and PRMT5 expression was measured by Western blot analysis (top) and qPCR (bottom). Data are the means ± SEM of three independent experiments. Statistical differences were determined by two-tailed unpaired Student’s t test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (B) Volcano plot showing the up- and downregulated genes in NCI-H460 cells between NC-shRNA and PRMT5-shRNA. Each group contained three replicates. (C) Bar graph of enriched terms of the differentially expressed genes between NC-shRNA and PRMT5-shRNA in HCI-H460 cells. Red boxes show the prominent GO terms. (D) Heatmap plot showing the negative regulation of T cell activation-associated genes between NCI-H460 PRMT5-shRNA and NC-shRNA cells. (E) Quantitative PCR verifying the candidate genes in NCI-H460 PRMT5-shRNA and NC-shRNA cells. The mean ± SEM is graphed. Statistical differences were determined by a two-tailed unpaired Student’s t test, *p < 0.05, **p < 0.01, ***p < 0.001. (F) PD-L1 expression was measured in NC-shRNA and PRMT5-shRNA NCI-H460 cells by flow cytometry. Data are representative of three independent experiments. Mean ± SEM was graphed. Statistical differences were determined by a two-tailed unpaired Student’s t test, **p < 0.01.