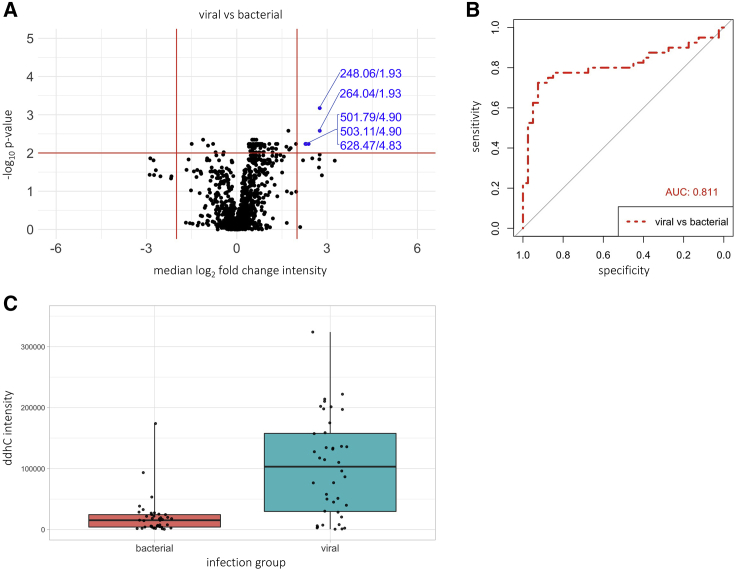
Figure 3.
ddhC differentiates viral versus bacterial infections in an independent validation cohort
Data from the validation cohort of sera from 40 viral and 40 bacterial infection patients undergoing the small molecule profiling assay. (A) volcano plot showing median log2 fold change in intensity of each feature versus -log10p value when comparing viral versus bacterial patients. Empirical threshold lines in red represent a fold-change of 4 [log2(fold-change) of 2] and p value of 0.01 [-log10(p value) of 2]. Features exceeding the threshold are shown in blue by mass:charge ratio/retention time, with 248.06/1.92 (ddhC) performing best. The next best performing feature 264.04/1.93 also corresponds to ddhC ([M+K]+ adduct). p-values generated using the two-sided Wilcoxon test and adjusted using the Benjamini-Hochberg procedure.
(B) Area under the receiver operating characteristic curve of 0.811 (95% CI 0.708-0.915) for ddhC discriminating between viral and bacterial infections.
(C) Relative ddhC intensity data in viral versus bacterial groups. Points represent individual patients. Boxes represent interquartile ranges with medians