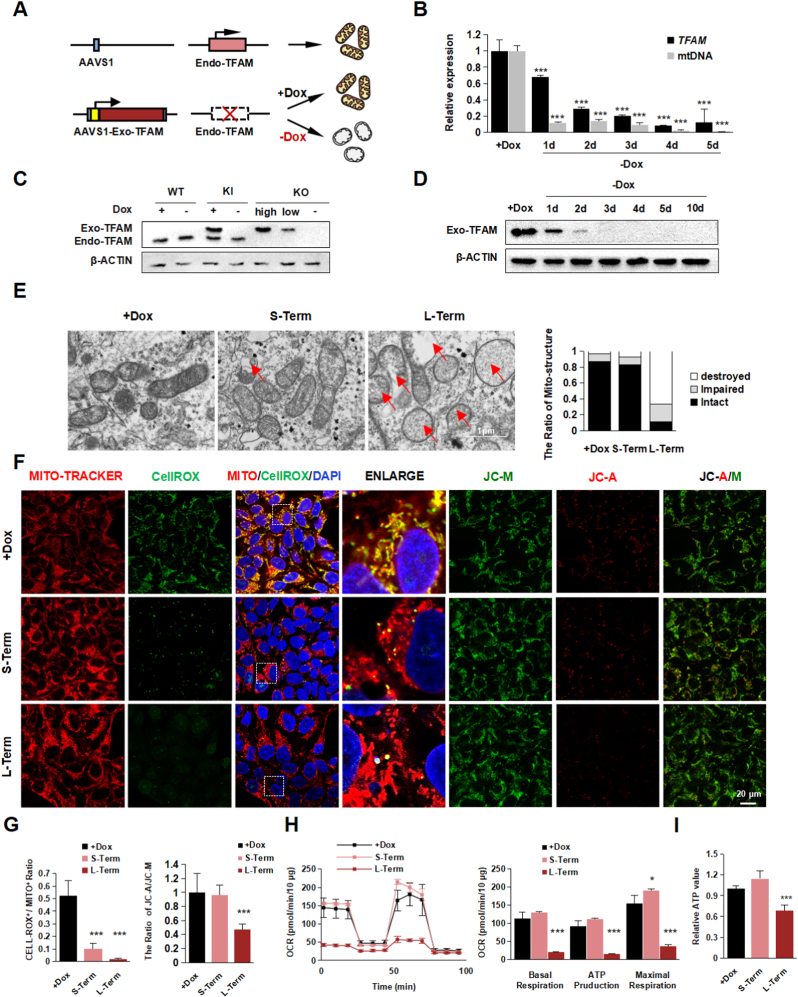
Fig. 1.
Establishment of TFAM-knockout hPSCs.
(A) Schematic diagram the procedure of generating Dox-induced TFAM-knockout hPSC line.
(B) Quantitative RT-PCR analysis of the expression level of TFAM and mtDNA in TFAM-/- hPSC after Dox withdrawal. Data were shown as Mean ± SEM (n = 3, ***p < 0.001 compared with the +Dox group).
(C) Western blot of the expression of Endo-TFAM and Exo-TFAM (3 × flag tagged) protein in WT, TFAMKI (Exo-TFAM knock-in), and TFAM-/- hPSC with or without Dox. β-actin served as the loading control.
(D) Western blot analysis of Exo-TFAM protein level in TFAM-/- hPSC after Dox removed. β-actin served as the loading control.
(E) Representative transmission electron photomicrographs showing the mitochondrial ultrastructure of TFAM-/- hPSCs treated with or without Dox. Quantification of intact, impaired, and destroyed mitochondria was calculated respectively. Data were shown as Mean ± SEM (n = 15).
(F–G) Representative immunofluorescence images (F) and the quantification (G) for CellRox (green)
/ Mito-tracker (red), DAPI (blue), and JC-A (red)/M (green) in Dox treated or untreated. Scale bars, 20 μm. Data were shown as Mean ± SEM (n = 7, ***p < 0.001 compared with the +Dox group).
(H) Seahorse assay of oxygen consumption rate (OCR) in TFAM-/- hPSCs treated with or without Dox and quantification of mitochondrial functional parameters. Data were shown as Mean ± SEM (n = 3, *p < 0.05, ***p < 0.001 compared with the +Dox group).
(I) The total ATP value in group +Dox, Short-Term, and Long-Term. Data were shown as Mean ± SEM (n = 3, ***p < 0.001 compared with the +Dox group). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)