Abstract
As an essential trace element, selenium (Se) plays a tremendous role in the functioning of the human organism being used for the biosynthesis of selenoproteins (proteins containing one or several selenocysteine residues). The functions of human selenoproteins in vivo are extremely diverse. Many selenoproteins have an antioxidant activity and, hence, play a key role in cell antioxidant defense and maintenance of redox homeostasis, which accounts for their involvement in diverse biological processes, such as signal transduction, proliferation, cell transformation and aging, ferroptosis, immune system functioning, etc. One of the critical functions of selenoenzymes is participation in the synthesis of thyroid hormones regulating basal metabolism in all body tissues. Over the last decades, optimization of population Se intake for prevention of diseases related to Se deficiency or excess has been recognized as a pressing issue in modern healthcare worldwide.
Keywords: selenium, selenocysteine, selenoproteins, antioxidants
INTRODUCTION
Selenium (Se) belongs to the essential trace elements necessary for normal human activity. Se enters the body with food of either plant or animal origin. Optimization of population Se intake for preventing disorders related to its deficiency or excess is one of the pressing issues in modern healthcare.
The necessity of Se for living organisms is due to the fact that it is a component of selenocysteine (Sec) found in various selenoproteins. Selenoproteins are synthesized in the cells via a unique mechanism that involves specific enzymes and factors and directly depends on Se intake. Human selenoproteome is encoded by 25 selenoprotein genes. The functions of the encoded proteins in the human body are extremely diverse. Many selenoproteins have a pronounced antioxidant activity, thereby playing a crucial role in the cell antioxidant defense, as well as maintenance of cell redox homeostasis. This activity accounts for their importance in multiple biological processes, such as signal transduction, proliferation, transformation, cell aging, ferroptosis, immune system activity, etc. Another important function of selenoenzymes is participation in the production of thyroid hormone regulating basal metabolism in virtually all body tissues.
This review discusses some aspects of Se metabolism and selenoprotein biosynthesis in the human body.
DISCOVERY OF SELENIUM, ITS PROPERTIES AND OCCURENCE
Se was discovered in 1817 by the Swedish chemist Jöns Jacob Berzelius. Due to the similarity of the new element to the previously discovered tellurium (from Latin tellus meaning Earth), Berzelius named it in 1782 in honor of the goddess of Moon (Greek selḗnē). Perhaps, a silver matte luster of the purified element also contributed to this decision [1].
In nature, Se exists as several crystalline modifications, as well in vitreous and amorphous forms. The oxidation states of Se are –2, 0, +4, +6. Se belongs to the sixth main group of the Periodic Table of Elements and displays both metallic and non-metallic properties. Along with oxygen, sulfur, tellurium, polonium, and artificially created radioactive livermorium, Se forms the so-called chalcogen group (from Greek word meaning ore-forming). Se, sulfur, and tellurium exhibit similar properties, but strongly differ in the relative occurrence in the Earth’s crust. In particular, the content of sulfur is by three orders of magnitude higher than the content of Se. Both elements are found in inorganic and organic worlds, in a free state, and in multiple compounds. Being chemically close elements, Se and sulfur form similar compounds and hold equivalent positions in many molecules [2-4].
Naturally occurring Se has six stable isotopes; the most common are 80Se (49.62%) and 78Se (23.51%). At least 16 more radioactive isotopes have been synthesized artificially by irradiating stable Se nuclei with neutrons. Among them, 75Se is widely used in biochemistry for examining metabolism of Se and selenoproteins [5].
Qualitative analysis of Se is currently performed rather rarely due to widespread distribution of this element. In particular, in the case of suspected Se poisoning, the element is identified in the exhaled air by gas chromatography [6]. Quantitative detection of Se is carried out by spectroscopic methods or neutron activation analysis [7, 8]. This allows to measure both total Se content in the samples and its content in certain compounds, which in turn, permits to analyze the metabolism of various Se-containing substances [3, 7-10].
Se is an extremely rare element, 59th most common in the Earth’s crust. Natural Se-containing compounds are mainly hydrogen selenide (H2Se) derivatives that exist in a mixture with sulfatides within copper-zinc, pyrite, copper-cobalt, and polymetallic ores. However, in a pure form, Se-containing minerals are found extremely rarely [1, 2, 4].
The average content of Se in the Earth’s crust is ~50 µg/kg. Se is distributed extremely heterogeneously in the soil, and its concentration in different geographic regions varies from 10 to 2000 µg/kg, averaging at ~400 µg/kg [3, 4]. The content of Se in the soil depends mainly on the parent material, climate, and use of fertilizers.
The average Se concentration in the world ocean is as low as 0.2 µg/liter, but in some regions, e.g., in hydrothermal vents (“black smokers”), the level of Se can be 50 to 100 times higher [11]. In inland lakes, the concentration of Se is ~25 µg/liter [12].
Se is also found in the atmosphere as a component of ash produced by the volcanic activity and burning of fossil fuels. The level of Se in the air near the Earth surface ranges from 0.1 to 10 ng/m3, whereas its local concentration in certain areas may be as high as 500 ng/m3 [13].
In living systems, Se can exist both as elementary Se and as a component of diverse chemical compounds, such as selenates, selenites, analogs of sulfur-containing amino acids (selenomethionine, Sec, methylselenocysteine, selenocystathionine), and proteins containing these amino acids. Plants are an essential source of Se both for human and animals. Plants are able to absorb and transform different (both inorganic and organic) forms of Se. The average content of Se in plants is within 0.01-10 mg/kg dry mass; its amount is determined by the type of soil and its pH value, amount of precipitation, temperature, and plant development phase [4, 14-17]. Both excess and deficiency of Se in the environment produce a negative effect on the plant growth and development.
SELENIUM METABOLISM
Se enters the human or animal body through the following chain: soil – plant – food. Hence, Se concentration in the human blood and tissues depends on the Se content in food, drinking water, plants, and soil. People absorb Se preferentially from solid food and accumulate up to 3-20 mg of this element, depending on the geographic region and traditional diet. Around 80% of Se is found in selenoproteins [18].
According to the FAO/WHO dietary recommendations, the intake of Se should comprise 26 µg/day for women and 34 µg/day for men. At the same time, at a dose of 150-200 µg/day, Se produces primarily the antioxidant and immunity-enhancing effects, as well as certain anticarcinogenic effect [19]. In Russia, the requirements for Se have been established at a level of 55 µg/day for women, 75 µg/day for men, and 10-50 µg/day for children [20]. In Germany, the recommended Se intake for adults ranges within 30-70 µg/day, but not over 300 µg/day [21]. In the USA, the recommended daily dietary allowance for Se is 55 µg/day for both men and women [22].
The actual values for the Se intake and its blood levels in some countries are shown in Table 1 [23-28].
Table 1.
Selenium intake and blood levels in different geographic regions worldwide
| Country | Daily intake (µg) | Blood/serum (µg/liter) |
|---|---|---|
| Russia | 15-130 | 67-106 |
| Japan | 27-89 | 80-155 |
| China* | 2-6990 | 5-7800 |
| USA | 60-160 | 100-350 |
| Canada | 113-220 | 143 |
| Germany | 38-47 | 89-98 |
| Tibet | 5-15 | 5-47 |
| Finland until 1984 | 40 | 69 |
| Finland after 1984 | 80 | 109 |
Note. Daily Se intake and its content in human serum markedly differ in different countries [23-28]. Introduction of Se-containing dietary supplements in some regions of Europe and Central Asia (Finland, Keshan County in China) with insufficient Se intake resulted in the long-term improvement of population health and disease prevention.
* Due to the heterogeneity of soil, Se intake and, subsequently, its concentration in the human serum differ greatly in different regions of China [28].
The content of Se in plant and animal food fluctuates significantly. In plants, it depends on the level of bioavailable Se in the soil. In animals, the content of Se depends on diet, Se level in the fodder, and use of mineral and vitamin supplements (i.e., indirectly depends on the quality of soil used for the fodder growth). Food supplements contain Se either as selenomethionine (SeMet) or sodium selenite/selenate. Se-rich fodder yeast have a multitude of Se-containing compounds beside SeMet; Se concentration in yeast may be as high as 1-2 mg/g dry weight [17].
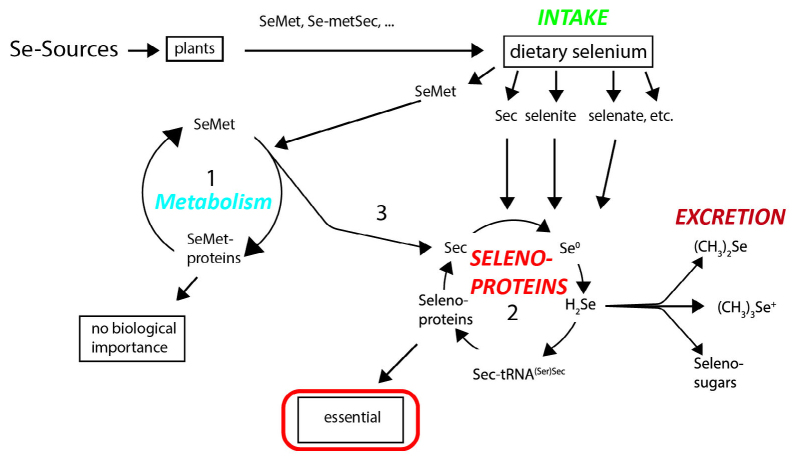
The chemical nature of consumed Se-containing compounds significantly affects Se absorption in human body (Fig. 1). Elementary Se, selenium dioxide, and selenium sulfide are poorly utilized, whereas selenites, selenates, and Se-containing amino acid derivatives are easily absorbed [29]. Absorption of Se-containing compounds takes place mainly in the duodenum and small intestine and, unlike absorption of other trace elements, seems to be independent of the Se status of the consumer [30]. Inorganic (selenite, selenate) and organic (Se-containing amino acids) compounds are absorbed with a comparable efficacy (70-95%), although via different transport mechanisms. Selenate is imported via the Na+/K+/Cl– cotransporter or OH– antiporter, selenite – via Na+-independent passive transport, whereas Se-containing amino acids are absorbed via specialized Na+-dependent amino acid transporters (similar to sulfur-containing amino acids) [31]. Further trafficking of Se compounds across the cell into the blood serum has not been fully elucidated [32]. Selenate ions and Se-Met enter the circulation mainly unchanged, whereas selenite is actively metabolized in the cells [33]. Sec and selenocystine penetrate into the circulation in small amounts. Most of these molecules bind to glutathione with the formation of mixed disulfides that are metabolized in the liver [34].
Fig. 1.
Metabolism of Se-containing compounds in human body. SeMet derived from plants is directly used for protein synthesis (1, SeMet cycle). Se released from SeMet and other compounds takes part in the Sec synthesis followed by the production of Sec-containing selenoproteins (2, selenoprotein cycle). H2Se is one of the intermediates in the generation of Sec-tRNA(Ser)Sec. Excessive Se is excreted from the body in the urine as trimethylselenonium ion and via respiratory tract in a form of dimethyl selenide. Se is also found in the urine in the content of selenosugars. Whether selenosugars take part in Se recirculation from the primary urine remains unknown.
Human cells produce three types of Se-containing proteins, relative amounts of which depend on the form of Se ingested with food (Fig. 1).
The first type are SeMet-containing proteins. SeMet is synthesized in plants and some fungi via sulfur assimilation pathway and enters human and animal body through the food. Then SeMet is recognized by methionine-specific aminoacyl-tRNA synthetase and incorporated into a nascent polypeptide chain similar to methionine. Protein biosynthesis in the presence of high SeMet concentrations results in a notably elevated content of this amino acid in the de novo synthesized proteins, e.g., serum albumin. Although more SeMet-containing proteins are produced as a result, this causes no overt biological effects. It is possible that SeMet-containing proteins serve as a Se depot, so that in the case of Se deficiency, some of them are mobilized for selenoprotein biosynthesis [34, 35], thereby ensuring normal body functioning.
The second type is represented by selenoproteins with Sec incorporated into the polypeptide chain at strictly defined positions via a specific mechanism (see below).
Finally, the third type includes poorly characterized Se-binding proteins that were identified experimentally using radioactive Se. The exact role of these proteins in Se metabolism remains largely unknown [36-39].
Se is removed from body in the exhaled air respiration as volatile dimethyl selenide formed by Se methylation [40] and in the urine as soluble trimethylselenonium ion [41]. Excretion of Se is significantly intensified in the case of excessive Se intake. Volatile dimethyl selenide accounts for typical unpleasant odor of the exhaled air in the case of Se intoxication. Normally, urine also contains selenosugars, namely 1-beta-methylseleno-N-acetyl-D-galactosamine and its precursor glutathione-seleno-N-acetyl-D-galactosamine [42]. There are data suggesting that these sugars are solely excretion products [43]. However, there is also evidence that selenosugars are involved in Se recirculation from the primary urine [44], but this hypothesis requires further investigation.
EFFECTS OF SELENIUM DEFICIENCY AND EXCESS ON HUMAN ORGANISM
Se is one of the essential trace elements that plays a critical role in numerous biological processes [37, 46-47]. Unlike other trace elements that may act as cofactors, active Se is always covalently bound to organic molecules. The biological role of Se is related to its presence in Sec, which is incorporated into specialized proteins called selenoproteins [48-50]. Optimization of Se intake by general human population poses one of the pressing issues of current healthcare system. A reasonable balance should be found, because, on one hand, Se is an essential dietary component, while on the other hand, it is a dangerous toxic substance [51-56].
In the early 20th century, the opinion of the world scientific community on Se has changed dramatically – from poison and carcinogen to one of the key essential trace elements with a paramount importance for human health [45, 57]. In 1980, the World Health Organization referred Se to essential dietary components.
Se deficiency. Pathologies associated with Se deficiency are rare, in particular, due to the wide use of Se-containing food supplements in the regions with low Se levels in the soil (Tibet, some regions of China) [58]. Se deficiency results in poor appetite, retarded body growth, and reduced total muscle mass. It also affects the functioning of the thyroid gland, cardiovascular and immune systems and may cause infertility, etc. [69-61]. Profound Se deficiency in a food chain results in the development of specific endemics, such as Keshan disease (cardiomyopathy) and Kashin–Beck disease (osteochondropathy) in humans, as well as myopathy and general muscular dystrophy in animals [62].
An essential role of Se in human health has been demonstrated in patients with muscular dystrophy. In particular, the symptoms of this disease were reversed in patients receiving Se supplements as parenteral nutrition (intravenous injections) [63]. Se-containing food supplements are currently considered as an integral part of parenteral nutrition [64, 65].
The pathological effects related to Se deficiency in vivo are caused by the decrease in the production of vital selenoproteins. The studies in animals have revealed that suppression of biosynthesis of certain selenoproteins or Se-specific transporters results in significant increase in the intracellular content of free radicals in cells followed by cell death and therefore, is incompatible with life [66, 67].
It should be noted that the extent of biosynthesis suppression upon Se deficiency is different for different selenoproteins, i.e., some of them are continuously produced, whereas the synthesis of others may be fully arrested. Vital organs, such as brain and endocrine glands, are supplied with Se even in the case of its deficiency due to specific regulatory mechanisms [55, 68, 69].
Se excess (selenosis). Symptoms of intoxication caused by excessive Se (selenosis) have been well studied in humans and animals. An extremely high dose of dietary Se ingested within a short time period causes acute toxicity rapidly resulting in death. However, consumption of moderately elevated Se amounts within longer time intervals causes chronic selenosis characterized by the body mass loss, hair loss, changes in the nail structure, dermatitis, disorders of the gastrointestinal tract, reduced fertility, and abnormalities in the offspring [70].
Selenosis was reported for the first time in the XIII century. When travelling in Western China, Marco Polo described nausea, softening of hooves, and loss of fur in his horses, which was due to the consumption of some local Se-rich plants. Cases of selenosis in domestic animals were also described in the second half of the XVI century in Columbia (South America) that were characterized by the massive hair loss, hoof and joint diseases, reproductive disorders, and even death. Moreover, people inhabiting that region were described to have developmental defects. Similar symptoms were observed in horses in the mid-XIX century in South Dakota (USA). Local doctors described the so-called alkali disease linked to the high soil salinity. Only in 1931, alkali disease was identified as chronic selenosis [71].
Selenosis is diagnosed quite rarely in medical practice. Statistical reports suggest that up to 80% of the population may experience dietary deficiency of Se, rather that its excess. If such diagnosis is established, the disease prognosis is typically positive, because selenosis can be reversed solely by correcting the diet [72, 73].
The toxicity of Se may be due to the non-specific replacement of sulfur with Se in the sulfur-containing amino acids and resulting alterations in the protein tertiary structure. Another hypothesis proposes that elevated Se concentrations disturb the redox balance in an organism with all the following consequences [73, 74].
SELENOPROTEIN BIOSYNTHESIS
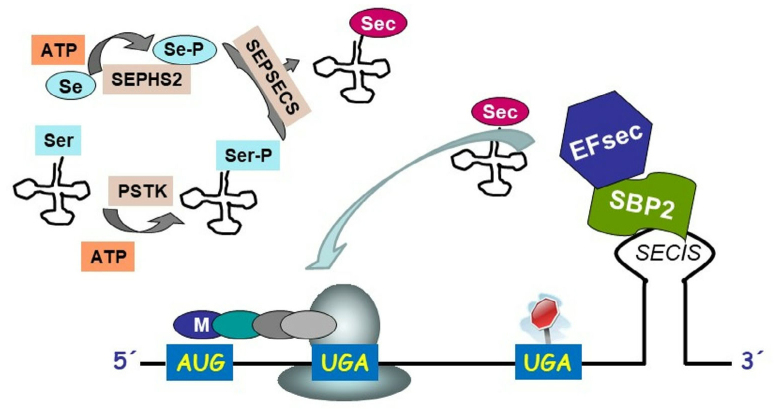
Selenoprotein biosynthesis is a complex process that proceeds in several stages. There is no free Sec in the body, and its synthesis occurs directly on specific tRNA (tRNA(Ser)Sec) with the UCA anticodon complementary to the UGA stop codon. First, tRNA(Ser)Sec is acylated with serine by seryl-tRNA synthetase with the formation of Ser-tRNA(Ser)Sec. The latter is not recognized by the common translation factors (bacterial EF-Tu and eukaryotic eEF1A) and cannot enter translation [48-50].
In eukaryotes and archaea, tRNA-bound serine residue is converted into Sec by three enzymes: O-phosphoseryl-tRNA [Ser] Sec kinase (PSTK), selenophosphate synthetase 2 (SEPHS2), and Sec synthase (SEPSECS). In particular, PSTK phosphorylates serine residue bound to tRNA, resulting in phosphoseryl-tRNA(Ser)Sec [75, 76], whereas SEPHS2 phosphorylates selenide to monoselenophosphate (HSeH2PO3) in the ATP-dependent manner. SEPHS2 contains its own Sec and, therefore, can participate in the regulation of selenoprotein biosynthesis [77]. Finally, SEPSECS catalyzes reaction between monoselenophosphate and phosphoseryl-tRNA(Ser)Sec by converting tRNA-bound phosphoserine into Sec, producing Sec-tRNA(Ser)Sec [78] (Fig. 2).
Fig. 2.
Selenoprotein biosynthesis. First, Sec-tRNA(Ser)Sec is produced in a three-stage process. Next, charged aminoacyl-tRNA in a complex with specific factors is transported to the free A-site of the ribosome that translates selenoprotein mRNA.
Selenocysteine is encoded by the UGA codon, which is normally one of the three stop codons necessary for terminating the elongation of the polypeptide chain. The UGA codon encodes selenocysteine only if the translated mRNA contains a unique selenocysteine insertion sequence (SECIS) element, a ~60 nucleotide motif that adopts a typical hairpin structure. SECIS can be recognized by the presence of particular nucleotides at specific positions, as well as the presence of non-canonical A-G base pairs. In bacteria, SECIS elements are located within the mRNA coding regions immediately after the UGA codon in the same reading frame. In archaea and eukaryotes, SECIS elements are located in the mRNA 3′-untranslated regions and may ensure incorporation of several Sec residues on different UGA codons [79-85].
Apart from the UGA codons and SECIS elements, biosynthesis of selenoproteins requires a set of protein factors, among which Sec-specific eukaryotic elongation factor EFsec and SECIS-binding protein 2 (SBP2) play a crucial role. SBP2 binds to SECIS and contains the RNA-binding domain similar to that found in the ribosomal protein L30. EFsec is homologous to the eukaryotic elongation factor EF-1A. Similar to this factor, EFsec mediates the delivery of aminoacyl-tRNA to the ribosome active site. Interestingly, in prokaryotes, the functions of EFsec and SBP2 are carried out by the two-domain SelB protein [79].
However, many details of the mechanism behind the biosynthesis of selenoproteins remain unclear. According to the current understanding (Fig. 2), EFsec forms a complex with Sec-tRNA(Ser)Sec (EFsec–Sec–tRNA(Ser)Sec), which binds to the SBP2–SECIS complex. Next, the formed multicomponent complex interacts with the ribosome translating the selenoprotein mRNA. This results in the delivery of charged Sec–tRNA(Ser)Sec to the free A-site of the ribosome, thereby accounting for the UGA-driven incorporation of Sec residue into the nascent polypeptide chain [79-85].
Biosynthesis of selenoproteins requires a set of additional factors, in particular ribosomal protein L30, nucleolin, and eukaryotic initiation factor eIF3a. Most likely, these proteins play an additional regulatory role in this process [86-88].
FUNCTIONS OF SELENOPROTEINS in vivo
Selenoproteins have been identified in the organisms from all three domains of life – eukaryotes, archaea, and bacteria. In eukaryotes, selenoproteins have been found in animals, but not in higher plants, yeast, or fungi. Currently, 25 genes encoding selenoproteins have been identified in humans (Table 2), 24 – in rodents, and 3 – in the fruit fly Drosophila melanogaster [89-91]. Moreover, at least 16 out of 25 human selenoproteins are enzymes containing Sec in the active site. Sec is a more acidic (pKa = 5.2 vs. 8.0) and nucleophilic residue than cysteine (Cys), and therefore, more catalytically active in the enzyme active site [48-50, 79].
Table 2.
Human selenoproteins
|
Antioxidant enzymes Redox signaling |
GPX1, GPX2, GPX3, GPX4, GPX6, SELENOK, SELENOR, SELENOW |
| TXNRD1, TXRND2, TXRND3 | |
| Thyroid hormone synthesis | DIO1, DIO2, DIO3 |
| Sec synthesis | Selenophosphate synthetase 2 (SEPHS2) |
| Se storage and transport | SELENOP |
| Protein folding | SELENOF, SELENON, SELENOM, SELENOS |
| Unknown functions | SELENOH, SELENOI, SELENOO, SELENOT, SELENOV |
Many selenoproteins have prominent antioxidant properties and can eliminate reactive oxygen species (ROS) formed as byproducts of molecular oxygen reactions in the process of oxidative phosphorylation in the cells. ROS can be also generated by the action of exogenous factors, such as drugs, xenobiotics, metals, radiation, smoking, and infection [92]. ROS include radical and non-radical oxygen forms produced by partial reduction of molecular oxygen, e.g., superoxide anionic radical (O2–), hydrogen peroxide (H2O2), and hydroxyl radical (OH•). At low concentration, ROS regulate many physiological processes, such as cell signaling, proliferation, tumor suppression, and immune system functioning. An increase in the ROS concentration results in the oxidative stress leading to the damage of nucleic acids, proteins, and lipids and causing numerous pathological processes, such as carcinogenesis [93], neurodegeneration [94, 95], atherosclerosis, diabetes [96], and aging [97]. Oxidative stress occurs as a result of imbalance between ROS generation and elimination.
Antioxidant selenoproteins play a pivotal role in the cell antioxidant defense, as well as the maintenance of redox homeostasis. This function accounts for their importance in the ROS-dependent biological processes, such as signal transduction, cell proliferation, transformation and aging, ferroptosis, immunity, and many others [49, 82, 84, 85].
Two major redox antioxidant systems in mammalian cells are glutathione and thioredoxin systems.
Glutathione peroxidases are a family of enzymes (GPX1-4, GPX6) catalyzing reduction of hydrogen peroxide to water and reduction of lipid hydroperoxides to the corresponding ethers. Glutathione peroxidases use glutathione as a cofactor that is then reduced by glutathione reductases. GPX1 was the first selenoprotein found in mammals [98].
Thioredoxin reductase is represented by three isoforms (TXNRD 1-3) containing Sec at the penultimate position in the polypeptide chain. TXNRD catalyzes reduction of thioredoxin and some other oxidized compounds, in particular, disulfide bonds in proteins [99, 100].
Iodothyronine deiodinases (DIOs) play a major role in the regulation of activity of thyroid hormones. The latter accelerate basal metabolism in the cells and act virtually on all body tissues. DIO1 and DIO2 convert inactive thyroxine (T4) to active triiodothyronine (T3), whereas DIO3 deactivates T4 or T3 by converting it to inactive rT3 or T2, thus playing a critical role in the regulation of body metabolism [45, 101, 102].
Selenophosphate synthetase 2 (SEPHS2) phosphorylates selenide into monoselenophosphate, which then plays an important role in selenoprotein biosynthesis (see above). Because this enzyme contains its own Sec, it can participate in the regulation of selenoprotein production [77].
Another protein that plays a key role in Se storage, transport and supply in vivo is selenoprotein P (SELENOP). This glycoprotein is found in the blood serum (3-6 mg/liter) and may account for up to 50% serum Se. The fundamental difference of SELENOP from other selenoproteins is that it contains not one, but several (up to 10 in humans) Sec residues per protein molecule. Along with Se transport, SELENOP can bind heavy metals and takes part in the organism defense against oxidative stress [103-105]. SELENOP serum concentration may be a marker for assessing body Se status, as well as overall body condition [106]. In particular, the level of SELENOP together with some other markers allows to predict the course of disease in COVID-19 patients [107], as well as predict and control recurrence of breast cancer [108].
Recently, it was found that some thyroiditis patients had pathogenic anti-SELENOP autoantibodies that negatively affect Se transport and thereby inhibit the activity of some selenoproteins [109].
Lately, it was found that some selenoproteins are present in the endoplasmic reticulum (ER), where they participate in protein folding and secretion, as well as ER stress response [110-112].
CONCLUSIONS
The essential trace element Se plays an extremely important role in the human organism and its functioning. Se is ingested with food of both plant and animal origin and utilized for the biosynthesis of functionally active selenoproteins that may contain one or several Sec residues. Se is a structural and functional cysteine analogue, in which the sulfur atom is replaced by a Se atom. Sec is the 21st proteinogenic amino acid, which is encoded by the UGA codon that otherwise serves as a stop codon in protein synthesis.
A significant progress has been achieved recently both in understanding the structure of selenoproteins and detailed molecular mechanisms underlying their biosynthesis. The functions of selenoproteins in the human body are extremely diverse. Many selenoproteins have a prominent antioxidant activity and play a key role in the cell antioxidant defense and maintenance of redox homeostasis, which accounts for their role in diverse biological processes, such as signal transduction, cell proliferation, transformation and ageing, ferroptosis, immune system function, etc. Involvement in the synthesis of thyroid hormones, which regulate basal metabolism in all body tissues, is another crucial function of selenoenzymes.
Over the last decades, optimization of populational Se intake for prevention of diseases related to Se deficiency or excess has been recognized as a pressing issue for the healthcare systems worldwide.
Acknowledgments
I express sincere appreciation to Lutz Schomburg and Kamil Demircan for critical comments and fruitful discussion. I would like to thank Irina Minich for assistance in the manuscript preparation. Also, I would like to take the opportunity to thank all my former colleagues for the wonderful time of collaboration during my work at the Laboratory of Protein Synthesis Regulation at the Institute of Protein Research, Pushchino, Russia.
Abbreviations
- Se
selenium
- Sec
selenocysteine
- SeMet
selenomethionine
Ethics declarations
The author declares no conflicts of interest. This article does not contain description of studies with the involvement of humans or animal subjects.
References
- 1.Flohé L., Andreesen J. R., Brigelius-Flohé R., Maiorino M., Ursini F. Selenium, the element of the moon, in life on earth. IUBMB Life. 2000;49:411–420. doi: 10.1080/152165400410263. [DOI] [PubMed] [Google Scholar]
- 2.Kabata-Pendias A. Geochemistry of selenium. J. Environ. Pathol. Toxicol. Oncol. 1998;17:173–177. [PubMed] [Google Scholar]
- 3.Fordyce F. M. Selenium geochemistry and health. Ambio. 2007;36:94–97. doi: 10.1579/0044-7447(2007)36[94:SGAH]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 4.Shamberger R. J. Selenium in the environment. Sci. Total Environ. 1981;17:59–74. doi: 10.1016/0048-9697(81)90108-X. [DOI] [PubMed] [Google Scholar]
- 5.Evenson J. K., Sunde R. A. Metabolism of tracer (75)Se selenium from inorganic and organic seleniumocompounds into selenoproteins in rats, and the missing (75)Se metabolites. Front. Nutr. 2021;8:699652. doi: 10.3389/fnut.2021.699652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uden P. C., Boakye H. T., Kahakachchi C., Tyson J. F. Selective detection and identification of Se containing compounds – review and recent developments. J. Chromatogr. A. 2004;1050:85–93. doi: 10.1016/j.chroma.2004.05.077. [DOI] [PubMed] [Google Scholar]
- 7.Fishbein L. Overview of analysis of carcinogenic and/or mutagenic metals in biological and environmental samples. I. Arsenic, beryllium, cadmium, chromium and selenium. Int. J. Environ. Anal. Chem. 1984;17:113–170. doi: 10.1080/03067318408079924. [DOI] [PubMed] [Google Scholar]
- 8.Weekley C. M., Aitken J. B., Finney L., Vogt S., Witting P. K., et al. Selenium metabolism in cancer cells: the combined application of XAS and XFM techniques to the problem of selenium speciation in biological systems. Nutrients. 2013;5:1734–1756. doi: 10.3390/nu5051734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki K. T., Ogra Y. Metabolic pathway for selenium in the body: speciation by HPLC-ICP MS with enriched Se. Food Additives Contam. 2002;19:974–983. doi: 10.1080/02652030210153578. [DOI] [PubMed] [Google Scholar]
- 10.Cappon C. J., Smith J. C. Mercury and selenium content and chemical form in fish muscle. Arch. Environ. Contam. Toxicol. 1981;10:305–319. doi: 10.1007/BF01055632. [DOI] [PubMed] [Google Scholar]
- 11.Rathgeber C., Yurkova N., Stackebrandt E., Beatty J. T., Yurkov V. Isolation of tellurite- and seleniumite-resistant bacteria from hydrothermal vents of the Juan de Fuca Ridge in the Pacific Ocean. Appl. Environ. Microbiol. 2002;68:4613–4622. doi: 10.1128/AEM.68.9.4613-4622.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crane M., Flower T., Holmes D., Watson S. The toxicity of selenium in experimental freshwater ponds. Arch. Environ. Contam. Toxicol. 1992;23:440–452. doi: 10.1007/BF00203807. [DOI] [PubMed] [Google Scholar]
- 13.Winkel L. H., Vriens B., Jones G. D., Schneider L. S., Pilon-Smits E., et al. Selenium cycling across soil-plant-atmosphere interfaces: a critical review. Nutrients. 2015;7:4199–4239. doi: 10.3390/nu7064199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pilon-Smits E. A. H. On the ecology of selenium accumulation in plants. Plants (Basel) 2019;8:197. doi: 10.3390/plants8070197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Z. Z., Wu L. Species richness and selenium accumulation of plants in soils with elevated concentration of selenium and salinity. Ecotoxicol. Environ. Safety. 1991;22:251–266. doi: 10.1016/0147-6513(91)90077-3. [DOI] [PubMed] [Google Scholar]
- 16.Terry N., Zayed A. M., De Souza M. P., Tarun A. S. Selenium in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000;51:401–432. doi: 10.1146/annurev.arplant.51.1.401. [DOI] [PubMed] [Google Scholar]
- 17.Schrauzer G. N. Selenomethionine: A review of its nutritional significance, metabolism and toxicity. J. Nutr. 2000;130:1653–1656. doi: 10.1093/jn/130.7.1653. [DOI] [PubMed] [Google Scholar]
- 18.Fan A. M., Book S. A., Neutra R. R., Epstein D. M. Selenium and human health implications in California’s San Joaquin Valley. J. Toxicol. Environ. Health. 1988;23:539–559. doi: 10.1080/15287398809531135. [DOI] [PubMed] [Google Scholar]
- 19.FAO, WHO (2002) Human Vitamin and Mineral Requirements. Report of a Joint FAO/WHO Expert Consultation, Bangkok, Thailand, FAO & WHO, Rome, 341 p.
- 20.Rospotrebnadzor (2009) Normal Range for Physiological Requirements in Energy and Nutrients for Various Groups of Population in the Russian Federation, Moscow, 36 p.
- 21.Hartfiel W., Bahners N. Selenium deficiency in the Federal Republic of Germany. Biol. Trace Element Res. 1988;15:1–12. doi: 10.1007/BF02990123. [DOI] [PubMed] [Google Scholar]
- 22.Food and Nutrition Board, Institute of Medicine (2000) Dietary Reference Intakes for Vitamin C, Vitamin E, Selenium, and Carotenoids: A Report of the Panel on Dietary Antioxidants and Related Compounds, Subcommitties on Upper Reference Levels of Nutrients and of Interpretation and Use of Dietary Reference Intakes, and the Standing Committee on the Scientific Evaluation of Dietary Reference Intakes, National Academy Press, Washington, 529 p.
- 23.Wu J., Xu G. L. Plasma selenium content, platelet glutathione peroxidase and superoxide dismutase activity of residents in Kashin-Beck disease affected area in China. J. Trace Elements Electrolytes Health Dis. 1987;1:39–43. [PubMed] [Google Scholar]
- 24.Alfthan G., Eurola M., Ekholm P., Venäläinen E. R., Root T., et al. Effects of nationwide addition of selenium to fertilizers on foods, and animal and human health in Finland: From deficiency to optimal selenium status of the population. J. Trace Elements Med. Biol. 2015;31:142–147. doi: 10.1016/j.jtemb.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 25.Aaseth J., Alexander J., Bjørklund G., Hestad K., Dusek P., et al. Treatment strategies in Alzheimer’s disease: a review with focus on selenium supplementation. Biometals. 2016;29:827–839. doi: 10.1007/s10534-016-9959-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stoffaneller R., Morse N. L. A review of dietary selenium intake and selenium status in Europe and the Middle East. Nutrients. 2015;7:1494–1537. doi: 10.3390/nu7031494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y., Rijntjes E., Wu Q., Lv H., Gao C., et al. Selenium deficiency is linearly associated with hypoglycemia in healthy adults. Redox Biol. 2020;37:101709. doi: 10.1016/j.redox.2020.101709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blazina T., Sun Y., Voegelin A., Lenz M., Berg M., et al. Terrestrial selenium distribution in China is potentially linked to monsoonal climate. Nat. Commun. 2014;5:4717. doi: 10.1038/ncomms5717. [DOI] [PubMed] [Google Scholar]
- 29.Wolffram S. Mechanisms of intestinal absorption of selenium. Med. Klin. (Munich) 1995;90 Suppl 1:1–5. [PubMed] [Google Scholar]
- 30.Ha H. Y., Alfulaij N., Berry M. J., Seale L. A. From selenium absorption to selenoprotein degradation. Biol. Trace Element Res. 2019;192:26–37. doi: 10.1007/s12011-019-01771-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato T., Read R., Rozga J., Burk R. F. Evidence for intestinal release of absorbed selenium in a form with high hepatic extraction. Am. J. Physiol. 1992;262:G854–858. doi: 10.1152/ajpgi.1992.262.5.G854. [DOI] [PubMed] [Google Scholar]
- 32.Whanger P., Vendeland S., Park Y. C., Xia Y. Metabolism of subtoxic levels of selenium in animals and humans. Ann. Clin. Lab. Sci. 1996;26:99–113. [PubMed] [Google Scholar]
- 33.Ferreira R. L. U., Sena-Evangelista K. C. M., de Azevedo E. P., Pinheiro F. I., Cobucci R. N., et al. Selenium in human health and gut microflora: Bioavailability of seleniumocompounds and relationship with diseases. Front. Nutr. 2021;8:685317. doi: 10.3389/fnut.2021.685317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hasegawa T., Mihara M., Okuno T., Nakamuro K., Sayato Y. Chemical form of selenium-containing metabolite in small intestine and liver of mice following orally administered seleniumocystine. Arch. Toxicol. 1995;69:312–317. doi: 10.1007/s002040050176. [DOI] [PubMed] [Google Scholar]
- 35.Burk R. F., Hill K. E., Motley A. K. Plasma selenium in specific and non-specific forms. Biofactors. 2001;14:107–114. doi: 10.1002/biof.5520140115. [DOI] [PubMed] [Google Scholar]
- 36.Köhrle J., Brigelius-Flohé R., Böck A., Gärtner R., Meyer O., et al. Selenium in biology: facts and medical perspectives. Biological Chemistry. 2000;381:849–864. doi: 10.1515/BC.2000.107. [DOI] [PubMed] [Google Scholar]
- 37.Bansal M. P., Mukhopadhyay T., Scott J., Cook R. G., Mukhopadhyay R., et al. DNA sequencing of a mouse liver protein that binds selenium: implications for selenium’s mechanism of action in cancer prevention. Carcinogenesis. 1990;11:2071–2073. doi: 10.1093/carcin/11.11.2071. [DOI] [PubMed] [Google Scholar]
- 38.Kühn E. C., Slagman A., Kühn-Heid E. C. D., Seelig J., Schwiebert C., et al. Circulating levels of selenium-binding protein 1 (SELENIUMBP1) are associated with risk for major adverse cardiac events and death. J. Trace Elements Med. Biol. 2019;52:247–253. doi: 10.1016/j.jtemb.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 39.Kühn-Heid E. C. D., Kühn E. C., Ney J., Wendt S., Seelig J., et al. Selenium-binding protein 1 indicates myocardial stress and risk for adverse outcome in cardiac surgery. Nutrients. 2019;11:2005. doi: 10.3390/nu11092005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hassoun B. S., Palmer I. S., Dwivedi C. Selenium detoxification by methylation. Res. Commun. Mol. Pathol. Pharmacol. 1995;90:133–142. [PubMed] [Google Scholar]
- 41.Nahapetian A. T., Janghorbani M., Young V. R. Urinary trimethylselenonium excretion by the rat: Effect of level and source of selenium-75. J. Nutr. 1983;113:401–411. doi: 10.1093/jn/113.2.401. [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi Y., Ogra Y., Ishiwata K., Takayama H., Aimi N., et al. Selenosugars are key and urinary metabolites for selenium excretion within the required to low-toxic range. Proc. Natl. Acad. Sci. USA. 2002;99:15932–15936. doi: 10.1073/pnas.252610699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burk R. F., Hill K. E., Motley A. K., Austin L. M., Norsworthy B. K. Deletion of selenoprotein P upregulates urinary selenium excretion and depresses whole-body selenium content. Biochim. Biophys. Acta. 2006;1760:1789–1793. doi: 10.1016/j.bbagen.2006.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki K. T., Somekawa L., Suzuki N. Distribution and reuse of 76Se-seleniumosugar in selenium-deficient rats. Toxicol Appl. Pharmacol. 2006;216:303–308. doi: 10.1016/j.taap.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 45.Köhrle J. The trace element selenium and the thyroid gland. Biochimie. 1999;81:527–533. doi: 10.1016/s0300-9084(99)80105-9. [DOI] [PubMed] [Google Scholar]
- 46.Fishbein L. Environmental selenium and its significance. Fundament. Appl. Toxicol. 1983;3:411–419. doi: 10.1016/s0272-0590(83)80014-1. [DOI] [PubMed] [Google Scholar]
- 47.Jackson M. L. Selenium: geochemical distribution and associations with human heart and cancer death rates and longevity in China and the United States. Biol. Trace Element Res. 1988;15:13–21. doi: 10.1007/BF02990124. [DOI] [PubMed] [Google Scholar]
- 48.Driscoll D. M., Copeland P. R. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 2003;23:17–40. doi: 10.1146/annurev.nutr.23.011702.073318. [DOI] [PubMed] [Google Scholar]
- 49.Schomburg, L. (2007) Molecular Regulation of Selenoprotein Biosynthesis and Selenium Transport, Doctoral Dissertation [in German], 1, 1-65.
- 50.Flohé L. The labour pains of biochemical seleniumology: the history of selenoprotein biosynthesis. Biochim. Biophys. Acta. 2009;1790:1389–1403. doi: 10.1016/j.bbagen.2009.03.031. [DOI] [PubMed] [Google Scholar]
- 51.Wilber C. G. Toxicology of selenium: A review. Clin. Toxicol. 1980;17:171–230. doi: 10.3109/15563658008985076. [DOI] [PubMed] [Google Scholar]
- 52.Barceloux D. G. Selenium. J. Toxicol. Clin. Toxicol. 1999;37:145–172. doi: 10.1081/CLT-100102417. [DOI] [PubMed] [Google Scholar]
- 53.Hu W., Zhao C., Hu H., Yin S. Food sources of selenium and its relationship with chronic diseases. Nutrients. 2021;13:1739. doi: 10.3390/nu13051739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winther K. H., Rayman M. P., Bonnema S. J., Hegedüs L. Selenium in thyroid disorders – essential knowledge for clinicians. Nat. Rev. Endocrinol. 2020;16:165–176. doi: 10.1038/s41574-019-0311-6. [DOI] [PubMed] [Google Scholar]
- 55.Schomburg L. The other view: the trace element selenium as a micronutrient in thyroid disease, diabetes, and beyond. Hormones (Athens) 2020;19:15–24. doi: 10.1007/s42000-019-00150-4. [DOI] [PubMed] [Google Scholar]
- 56.Schwarz K., Foltz C. M. Selenium as an integral part of Factor 3 against dietary necrotic liver degeneration. J. Am. Chem. Soc. 1957;79:3292–3293. doi: 10.1021/ja01569a087. [DOI] [PubMed] [Google Scholar]
- 57.Vernie L. N. Selenium in carcinogenesis. Biochim. Biophys. Acta. 1984;738:203–217. doi: 10.1016/0304-419X(83)90004-5. [DOI] [PubMed] [Google Scholar]
- 58.Alfthan G., Xu G. L., Tan W. H., Aro A., Wu J., et al. Selenium supplementation of children in a selenium-deficient area in China: Blood selenium levels and glutathione peroxidase activities. Biol. Trace Element Res. 2000;73:113–125. doi: 10.1385/BTER:73:2:113. [DOI] [PubMed] [Google Scholar]
- 59.Rayman M. P. The importance of selenium to human health. Lancet. 2000;356:233–241. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- 60.Brown K. M., Arthur J. R. Selenium, selenoproteins and human health: A review. Public Health Nutr. 2001;4:593–599. doi: 10.1079/PHN2001143. [DOI] [PubMed] [Google Scholar]
- 61.Koller L. D., Exon J. H. The two faces of selenium-deficiency and toxicity – are similar in animals and man. Can. J. Veter. Res. 1986;50:297–306. [PMC free article] [PubMed] [Google Scholar]
- 62.Combs, S. B. (1986) The Role of Selenium in Nutrition (Combs, S. B. and Combs, J. F., eds.) Academic Press Inc., Orlando, FL, 532 p.
- 63.Van Rij A. M., Thomson C. D., McKenzie J. M., Robinson M. F. Selenium deficiency in total parenteral nutrition. Am. J. Clin. Nutr. 1979;32:2076–2085. doi: 10.1093/ajcn/32.10.2076. [DOI] [PubMed] [Google Scholar]
- 64.Foster L. H., Sumar S. Selenium in health and disease: A review. Crit. Rev. Food Sci. Nutr. 1997;37:211–228. doi: 10.1080/10408399709527773. [DOI] [PubMed] [Google Scholar]
- 65.Rao A., Jericho H., Patton T., Sriram S., Hebert T., et al. Factors affecting selenium status in infants on parenteral nutrition therapy. J. Pediatr. Gastroenterol. Nutr. 2021;73:e73–e78. doi: 10.1097/MPG.0000000000003174. [DOI] [PubMed] [Google Scholar]
- 66.Schweizer U., Schomburg L. New insights into the physiological actions of selenoproteins from genetically modified mice. IUBMB Life. 2005;57:737–744. doi: 10.1080/15216540500364255. [DOI] [PubMed] [Google Scholar]
- 67.Conrad M., Schweizer U. Unveiling the molecular mechanisms behind selenium-related diseases through knockout mouse studies. Antioxid. Redox Signal. 2010;12:851–865. doi: 10.1089/ars.2009.2912. [DOI] [PubMed] [Google Scholar]
- 68.Gross M., Oertel M., Köhrle J. Differential selenium-dependent expression of type I 5′-deiodinase and glutathione peroxidase in the porcine epithelial kidney cell line LLC-PK1. Biochem. J. 1995;306:851–856. doi: 10.1042/bj3060851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bermano G., Nicol F., Dyer J. A., Sunde R. A., Beckett G. J., et al. Tissue-specific regulation of selenoenzyme gene expression during selenium deficiency in rats. Biochem. J. 1995;311:425–430. doi: 10.1042/bj3110425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tinggi U. Essentiality and toxicity of selenium and its status in Australia: a review. Toxicol. Lett. 2003;137:103–110. doi: 10.1016/s0378-4274(02)00384-3. [DOI] [PubMed] [Google Scholar]
- 71.Rosenfeld, I. (1964) Selenium: Geobotany, Biochemistry, Toxicity and Nitrition (Rosenfeld, I., and Beath, O. A., eds.) Academic Press, New York, 411 p., 10.1016/C2013-0-12047-8.
- 72.Maier, K. J., and Knight, A. W. (1994) Ecotoxicology of selenium in freshwater systems, in Reviews of Environmental Contamination and Toxicology (Ware, G. W., ed.) Vol. 134. Springer, New York, NY, pp. 31-48, 10.1007/978-1-4684-7068-0_2. [DOI] [PubMed]
- 73.Sunde R. A. The biochemistry of selenoproteins. J. Am. Chem. Soc. 1984;61:1891–1900. doi: 10.1007/BF02540827. [DOI] [Google Scholar]
- 74.Spallholz J. E., Hoffman D. J. Selenium toxicity: Cause and effects in aquatic birds. Aquatic Toxicol. 2002;57:27–37. doi: 10.1016/s0166-445x(01)00268-5. [DOI] [PubMed] [Google Scholar]
- 75.Yuan J., Palioura S., Salazar J. C., Su D., O’Donoghue P., et al. RNA-dependent conversion of phosphoserine forms seleniumocysteine in eukaryotes and archaea. Proc. Natl. Acad. Sci. USA. 2006;103:18923–18927. doi: 10.1073/pnas.0609703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu X. M., Carlson B. A., Mix H., Zhang Y., Saira K., et al. Biosynthesis of seleniumocysteine on its tRNA in eukaryates. PLoS Biol. 2007;5:e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guimarães M. D., Peterson D., Vicari A., Cocks B. G., Copeland N. G., et al. Identification of a novel selD homolog from eukaryotes, bacteria, and archaea: is there an autoregulatory mechanism in seleniumocysteine metabolism? Proc. Natl. Acad. Sci. USA. 1996;93:15086–15091. doi: 10.1073/pnas.93.26.15086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Böck A., Forchhammer K., Heider J., Baron C. Selenoprotein synthesis: an expansion of the genetic code. Trends Biochem. Sci. 1991;16:463–467. doi: 10.1016/0968-0004(91)90180-4. [DOI] [PubMed] [Google Scholar]
- 79.Böck A., Forchhammer K., Heider J., Leinfelder W., Sawers G., et al. Selenocysteine: the 21st amino acid. Mol. Microbiol. 1991;5:515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- 80.Berry M. J., Banu L., Chen Y. Y., Mandel S. J., Kieffer J. D., et al. Recognition of UGA as a seleniumocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature. 1991;353:273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 81.Krol A. Evolutionarily different RNA motifs and RNA-protein complexes to achieve selenoprotein synthesis. Biochimie. 2002;84:765–774. doi: 10.1016/s0300-9084(02)01405-0. [DOI] [PubMed] [Google Scholar]
- 82.Papp L. V., Lu J., Holmgren A., Khanna K. K. From selenium to selenoproteins: Synthesis, identity, and their role in human health. Antioxid. Redox Signal. 2007;9:775–806. doi: 10.1089/ars.2007.1528. [DOI] [PubMed] [Google Scholar]
- 83.Seeher S., Mahdi Y., Schweizer U. Post-transcriptional control of selenoprotein biosynthesis. Curr. Protein Pept. Sci. 2012;13:337–346. doi: 10.2174/138920312801619448. [DOI] [PubMed] [Google Scholar]
- 84.Lu J., Holgren A. Selenoproteins. J. Biol. Chem. 2009;284:723–727. doi: 10.1074/jbc.R800045200. [DOI] [PubMed] [Google Scholar]
- 85.Hatfield D. L., Tsuji P. A., Carlson B. A., Gladyshev V. N. Selenium and seleniumocysteine: Roles in cancer, health, and development. Trends Biochem. Sci. 2014;39:112–120. doi: 10.1016/j.tibs.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chavatte L., Brown B. A., Driscoll D. M. Ribosomal protein L30 is a component of the UGA-seleniumocysteine recoding machinery in eukaryotes. Nat. Struct. Mol. Biol. 2005;12:408–416. doi: 10.1038/nsmb922. [DOI] [PubMed] [Google Scholar]
- 87.Miniard A. C., Middleton L. M., Budiman M. E., Gerber C. A., Driscoll D. M. Nucleolin binds to a subset of selenoprotein mRNAs and regulates their expression. Nucleic Acids Res. 2010;38:4807–4820. doi: 10.1093/nar/gkq247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Budiman M. E., Bubenik J. L., Miniard A. C., Middleton L. M., Gerber C. A., et al. Eukaryotic initiation factor 4a3 is a selenium-regulated RNA-binding protein that selectively inhibits selenocysteine incorporation. Mol. Cell. 2009;35:479–489. doi: 10.1016/j.molcel.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hatfield D. L., Gladyshev V. N. How selenium has altered our understanding of the genetic code. Mol. Cell. Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kryukov G. V., Castellano S., Novoselov S. V., Lobanov A. V., Zehtab O., et al. Characterization of mammalian seleniumoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 91.Gladyshev V. N., Arnér E. S., Berry M. J., Brigelius-Flohé R., Bruford E. A., et al. Selenoprotein gene nomenclature. J. Biol. Chem. 2016;291:24036–24040. doi: 10.1074/jbc.M116.756155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ray P. D., Huang B. W., Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Trachootham D., Alexandre J., Huang P. Targeting cancer cells by ROS-mediated mechanisms: A radical therapeutic approach? Nat. Rev. Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 94.Andersen J. K. Oxidative stress in neurodegeneration: Cause or consequence? Nat. Med. 2004;10:S18–S25. doi: 10.1038/nrn1434. [DOI] [PubMed] [Google Scholar]
- 95.Shukla V., Mishra S. K., Pant H. C. Oxidative stress in neurodegeneration. Adv. Pharmacol. Sci. 2011;2011:572634. doi: 10.1155/2011/572634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Paravicini T. M., Touyz R. M. Redox signaling in hypertension. Cardiovasc. Res. 2006;71:247–258. doi: 10.1016/j.cardiores.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 97.Haigis M. C., Yankner B. A. The aging stress response. Mol. Cell. 2010;40:333–344. doi: 10.1016/j.molcel.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brigelius-Flohé R., Maiorino M. Glutathione peroxidases. Biochim. Biophys. Acta. 2012;1830:3289–3303. doi: 10.1016/j.bbagen.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 99.Arnér E. S. Focus on mammalian thioredoxin reductases – important selenoproteins with versatile functions. Biochim. Biophys. Acta. 2009;1790:495–526. doi: 10.1016/j.bbagen.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 100.Arnér E. S., Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 101.Schomburg L., Köhrle J. On the importance of selenium and iodine metabo-lism for thyroid hormone biosynthesis and human health. Mol. Nutr. Food Res. 2008;52:1235–1246. doi: 10.1002/mnfr.200700465. [DOI] [PubMed] [Google Scholar]
- 102.Köhrle J., Jakob F., Contempré B., Dumont J. E. Selenium, the thyroid, and the endocrine system. Endocr. Rev. 2005;26:944–984. doi: 10.1210/er.2001-0034. [DOI] [PubMed] [Google Scholar]
- 103.Åkesson B., Bellew T., Burk R. F. Purification of selenoprotein P from human plasma. Biochim. Biophys. Acta. 1994;1204:243–249. doi: 10.1016/0167-4838(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 104.Burk R. F., Hill K. E. Selenoprotein P: A selenium richextracellular glycoprotein. J. Nutr. 1994;124:1891–1897. doi: 10.1093/jn/124.10.1891. [DOI] [PubMed] [Google Scholar]
- 105.Lamarche J., Ronga L., Szpunar J., Lobinski R. Characterization and quantification of selenoprotein P: Challenges to mass spectrometry. Int. J. Mol. Sci. 2021;22:6283. doi: 10.3390/ijms22126283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hybsier S., Schulz T., Wu Z., Demuth I., Minich W. B., et al. Sex-specific and inter-individual differences in biomarkers of selenium status identified by a calibrated ELISA for selenoprotein P. Redox Biol. 2017;11:403–414. doi: 10.1016/j.redox.2016.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Heller R. A., Sun Q., Hackler J., Seelig J., Seibert L., et al. Prediction of survival odds in COVID-19 by zinc, age and selenoprotein P as composite biomarker. Redox Biol. 2021;38:101764. doi: 10.1016/j.redox.2020.101764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Demircan K., Bengtsson Y., Sun Q., Brange A., Vallon-Christersson J., et al. Serum selenium, selenoprotein P and glutathione peroxidase 3 as predictors of mortality and recurrence following breast cancer diagnosis: A multicentre cohort study. Redox Biol. 2021;47:102145. doi: 10.1016/j.redox.2021.102145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sun Q., Mehl S., Renko K., Seemann P., Gorlich C. L., et al. Natural autoimmunity to selenoprotein p impairs selenium transport in Hashimoto’s thyroiditis. Int. J. Mol. Sci. 2021;22:13088. doi: 10.3390/ijms222313088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rocca C., Pasqua T., Boukhzar L., Anouar Y., Angelone T. Progress in the emerging role of selenoproteins in cardiovascular disease: Focus on endoplasmic reticulum-resident selenoproteins. Cell. Mol. Life Sci. 2019;76:3969–3985. doi: 10.1007/s00018-019-03195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pitts M. W., Hoffmann P. R. Endoplasmic reticulum-resident selenoproteins as regulators of calcium signaling and homeostasis. Cell Calcium. 2018;70:76–86. doi: 10.1016/j.ceca.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Addinsall A. B., Wright C. R., Andrikopoulos S., van der Poel C., Stupka N. Emerging roles of endoplasmic reticulum-resident selenoproteins in the regulation of cellular stress responses and the implications for metabolic disease. Biochem. J. 2018;475:1037–1057. doi: 10.1042/BCJ20170920. [DOI] [PubMed] [Google Scholar]