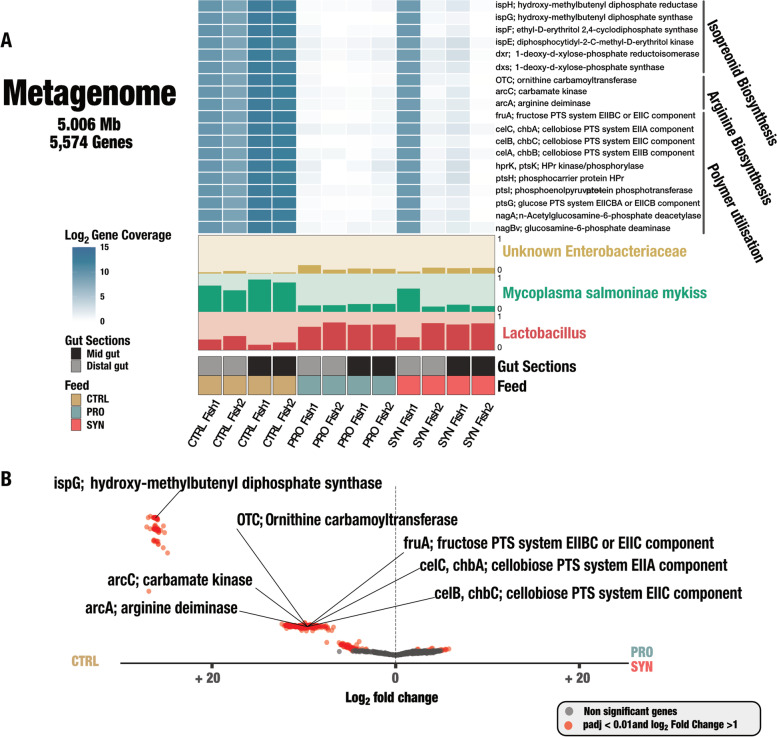
Fig. 4.
Genome-resolved metagenomics from gut microbiome across feeding type and intestinal sections. A Three metagenome-assembled genomes (MAGs) were resolved from the metagenome, consisting of 5.006 megabases (Mb) and 5574 non-redundant genes, which were visualised as yellow for Unknown Enterobacteriaceae, green for Candidatus Mycoplasma salmoninae mykiss, and red for Unknown Lactobacillus. Barplots indicate relative abundance of the two MAGs within each sample. Grouping of rainbow trout reared on different feeding types were visualised as orange for CTRL, blue for PRO, and red for SYN. Intestinal sections (gut sections) are coloured as black for mid gut and grey for distal gut. Heatmap visualises a series of genes of interest, which are related to isoprenoid biosynthesis, arginine biosynthesis, and polymer utilisation. Intensity of blue colour indicates log10 of coverage of genes across samples. B Volcano plot of differentially abundant metagenomic genes between samples from CTRL vs. PRO and SYN. Colouration of nodes indicate significance, whereas red nodes are significant genes after correction and have a log2 fold change (FC) > 1. Grey nodes are non-significantly differentiated genes between feeding types.