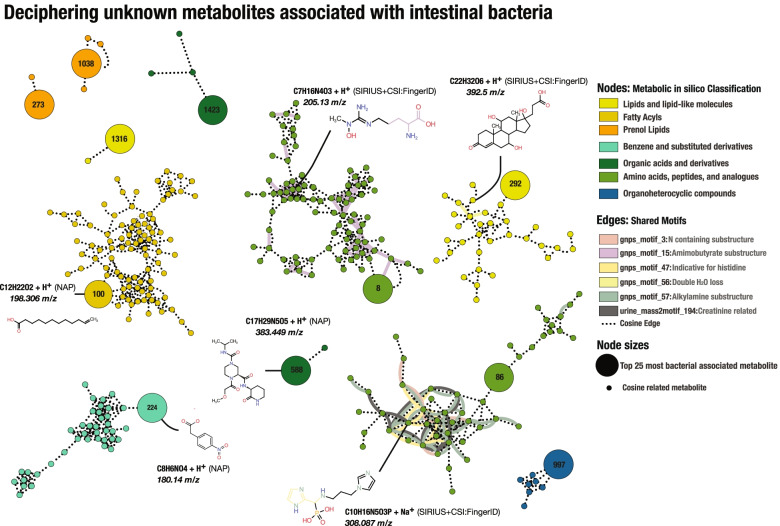
Fig. 6.
Molecular network showing bacterial associated metabolites and their molecular families with in silico annotation and hypothetical structures. Metabolomics-based molecular network of 11 bacterial associated metabolites (BAMs) and their molecular families coloured by putative chemical classes retrieved through a consensus of SIRIUS+CSI:FingerID and the GNPS based MolNetEnhancer workflow as indicated in the legend. A total of 350 metabolites were selected based on the bacterial association test and cosine relation to BAMs. Neighbour nodes of the significant nodes were co-selected to increase knowledge on chemical structural information of BAMs. Small nodes indicate metabolites in a network with bacterial associated metabolites. Edges are coloured according to shared motifs from MS2LDA and MotifDB databases or spectral similarity (cosine score > 0.7) as indicated in the legend. Unclassified molecular families were removed from the figure to increase clarity. Hypothetical structures suggested by in silico are shown and substructural motifs are highlighted with corresponding edge colours in the showed hypothetical structures