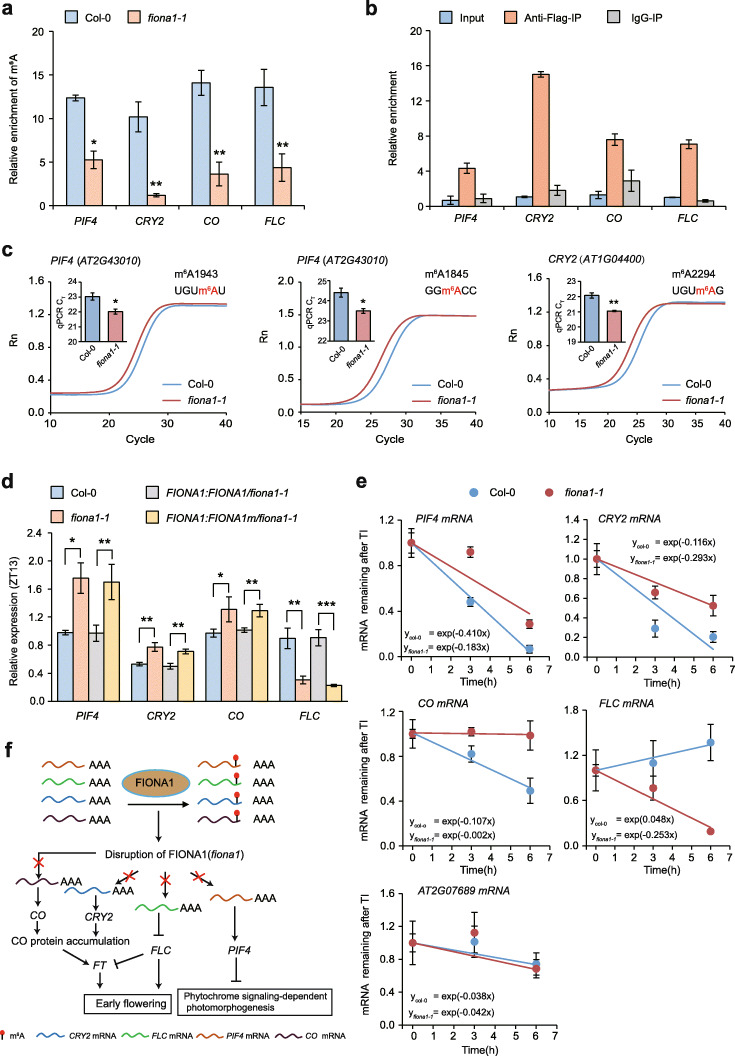
Fig. 6.
FIONA1-mediated m6A installation affects the stability of PIF4, CRY2, and FLC transcripts. a m6A-IP-qPCR results showing the relative m6A levels of PIF4, CRY2, and FLC transcripts in 12-day-old Col-0 and fiona1-1 seedlings. Data are means ± SD for 3 biological replicates × 3 technical replicates. * p < 0.05, ** p < 0.01 by t test (two-tailed). b RIP-qPCR assays in FIONA1:FIONA1-FLAG/ fiona1-1 plants showing that FIONA1 directly binds PIF4, CRY2, and FLC transcripts. Data are means ± SD for 3 biological replicates × 2 technical replicates. c Real-time fluorescence amplification curves and bar plot of the threshold cycle (CT) of qPCR showing SELECT results for detecting the FIONA1-targeted m6A sites in PIF4 and CRY2 mRNAs in Col-0 and fiona1-1 seedlings. Rn is the raw fluorescence for the associated well normalized to the fluorescence of the passive reference dye (ROX). Data for bar plot are means ± SD for 3 biological replicates × 2 technical replicates. * p < 0.05, ** p < 0.01 by t test (two-tailed). d The relative expression levels of PIF4, CRY2 and FLC at ZT13 in the indicated genotypic plants under LD conditions. TUB2 was used as the internal control gene. Data are means ± SD for 3 biological replicates × 3 technical replicates. * p < 0.05, ** p < 0.01 by t test (two-tailed). e The mRNA lifetimes of PIF4, CRY2, and FLC in Col-0 and fiona1-1. The AT2G07689 was used as the negative control. TI: transcription inhibition. Data are represented as means ± SD for 2 biological replicates × 3 technical replicates. f Proposed model describing the molecular mechanism through which FIONA1-mediated m6A installation regulates Arabidopsis phytochrome signaling-dependent photomorphogenesis and floral transition