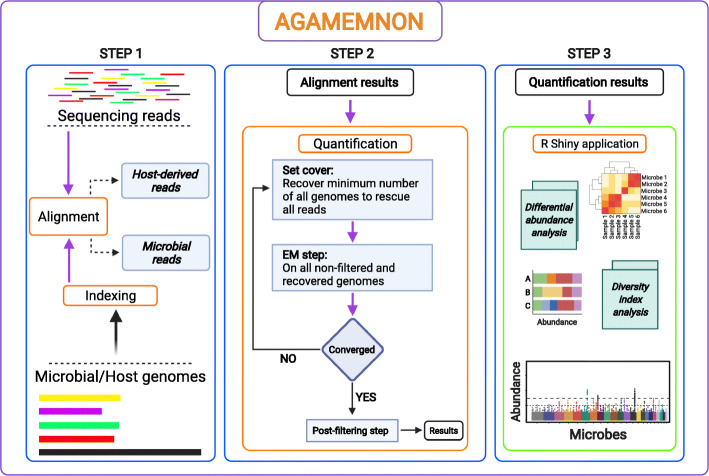
Fig. 1.
Schematic representation of AGAMEMNON. Dataset input is in raw FASTQ format. Paired-end (PE) or Single-end (SE) libraries are supported. For single-cell libraries, AGAMEMNON has helper scripts to enable per-cell analyses. In case of host tissue samples or contaminant quantification activities, the reads are first aligned against the host genome and the contaminant reference index using HISAT2. The host alignment file is saved for downstream applications and the resulting unmapped reads are forwarded to the main metagenomics/metatranscriptomics pipeline. Selective alignment is performed on the microbial reads against the reference index, while microbial abundances are subsequently quantified. A raw quantification table is produced as well as a taxonomic rank table. The results of the analysis can be used as input to AGAMEMNON’s R-Shiny application, which enables diverse analyses and investigations from a graphic user interface, including visualizations, dimensionality reduction, differential abundance, and diversity index analyses