Figure 4. Fbxw7 is necessary for tumor-intrinsic type I interferon signaling.
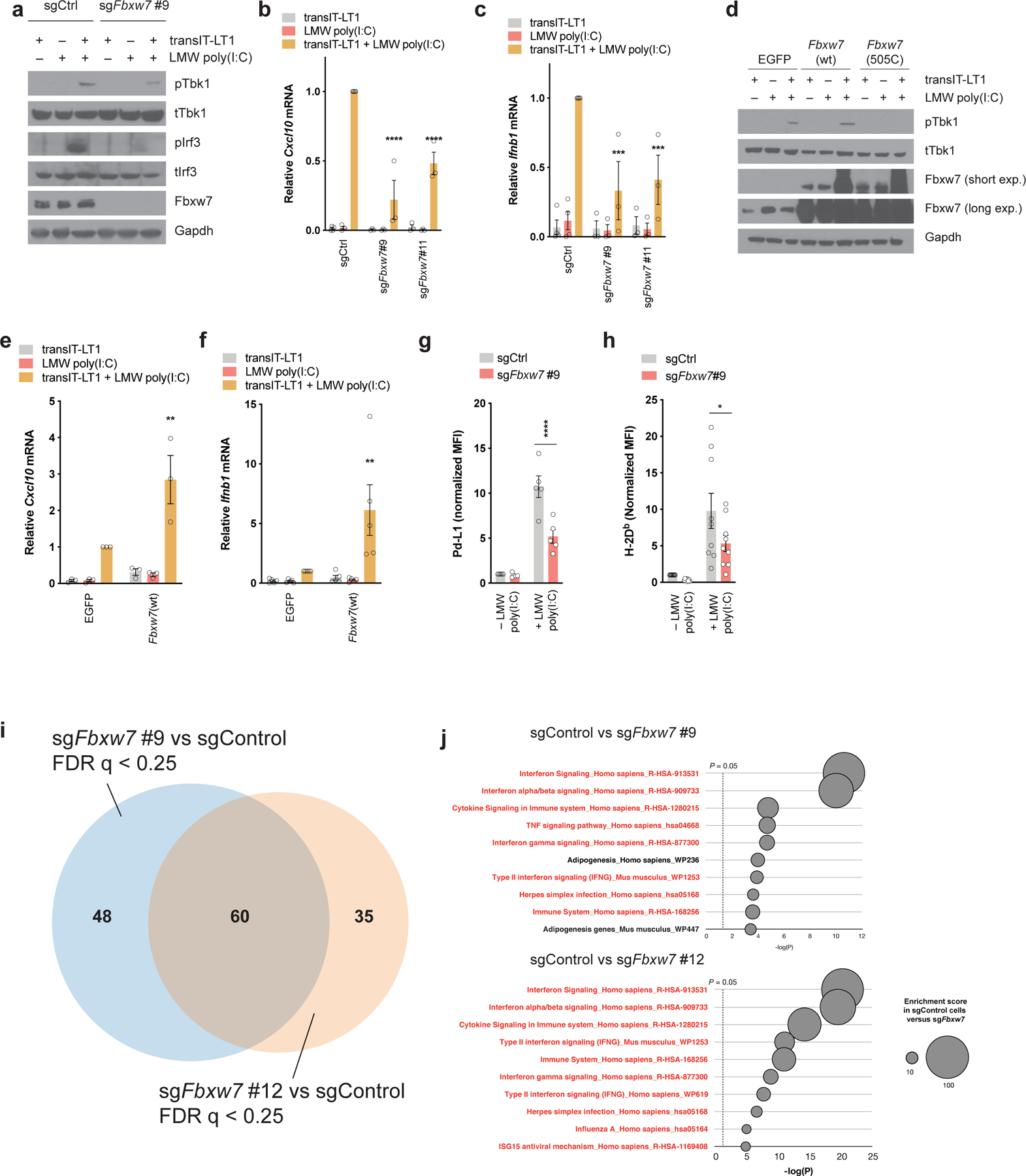
(a) Western blot of Tbk1/Irf3 signaling pathway in control and Fbxw7-deficient D4C9 cells following transfection with LMW poly(I:C) (1μg/ml) for 24h. (b-c) Relative (b) Cxcl10 and (c) Ifnb1 mRNA expression in control and Fbxw7-deficient D4C9 cells following transfection with LMW poly(I:C) (1μg/ml) for 24h. Each dot represents a biological replicate. Two-way ANOVA with Sidak’s multiple comparisons test was used to determine statistical significance, in comparison to D4C9-sgCtrl + transIT-LT1 + LMW poly (I:C). ***, P <0.001; ****, P <0.0001. (d) Western blot of Tbk1 activation in D4C9 cells overexpressing Egfp, wild-type Fbxw7 or Fbxw7(R505C) following transfection with LMW poly(I:C) (1μg/ml) for 24h. (e-f) Relative (e) Cxcl10 and (f) Ifnb1 mRNA expression in D4C9 cells overexpressing Egfp or wild-type Fbxw7, following transfection with LMW poly(I:C) (1μg/ml) for 24h. Each dot represents a biological replicate. Two-way ANOVA with Sidak’s multiple comparisons test was used to determine statistical significance, in comparison to D4C9-EGFP + transIT-LT1 + LMW poly (I:C). **, P <0.01. (g-h) Expression of (g) Pd-L1 and (h) MHC-I (H-2Db) in control and Fbxw7-deficient D4C9 cells following transfection with LMW poly(I:C) (1μg/ml) for 24h. MFI (median fluorescent intensity) was measured relative to sgCtrl. Each dot represents a biological replicate. Two-way ANOVA with Sidak’s multiple comparisons test was used to determine statistical significance. *, P <0.05; ****, P <0.0001. (i-j) RNA-seq analysis of Fbxw7-deficient and control D4C9 cells treated with Ifnγ (10ng/ml) for 24h. (i) Venn diagram showing significantly altered genes sets (false discovery rate q < 0.25) in sgFbxw7#9 and sgFbxw7#12 cells compared to control cells in presence of Ifnγ. (j) Most significantly downregulated gene sets in sgFbxw7#9 and sgFbxw7#12 cells compared to control cells in presence of Ifnγ. Highlighted in red are the gene sets related to viral sensing and interferon signaling.
