FIGURE 2.
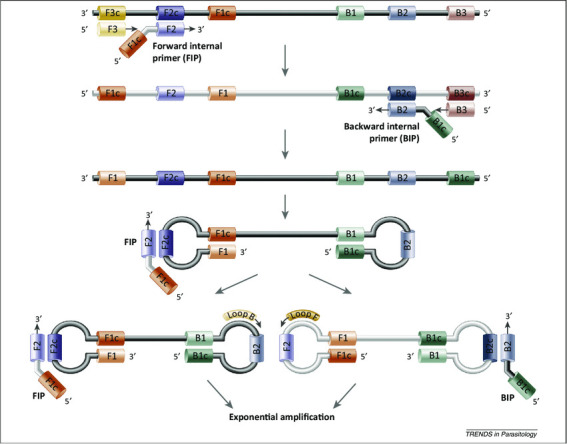
LAMP mechanism. The LAMPs employ 2 sets of primers, forward/backward internal primers (FIP and BIP) and outer primers (F3 and B3) to target 6 distinct regions (F1c, F2c, F3c sites on 1 end, and B1, B2, B3 sites on the other). The reaction is initiated by the binding of FIP to the F2c region on the double-stranded DNA. As the polymerase elongates the DNA from the FIP, the outer primer F3, which is shorter in length and lower in concentration than the FIP, binds onto its complementary region on the DNA and starts to displace the newly synthesized DNA. The replaced strand then forms a loop structure at one end because of the complementarity of F1 and F1c. This results in a single-stranded, double-stem-loop DNA structure (the so-called “dumb-bell” structure) with similar performance for BIP and B3. This dumbbell-structured DNA enters the amplification cycle because it is already self-primed. Elongation by the polymerase can occur from the free 3′-end of the single-stranded DNA (ssDNA) and from binding of the FIP/BIP primers to the single-stranded loop or from the optional accelerating loop primers (see Supplemental Fig. S1, with permission from Alhassan et al. 2015).
