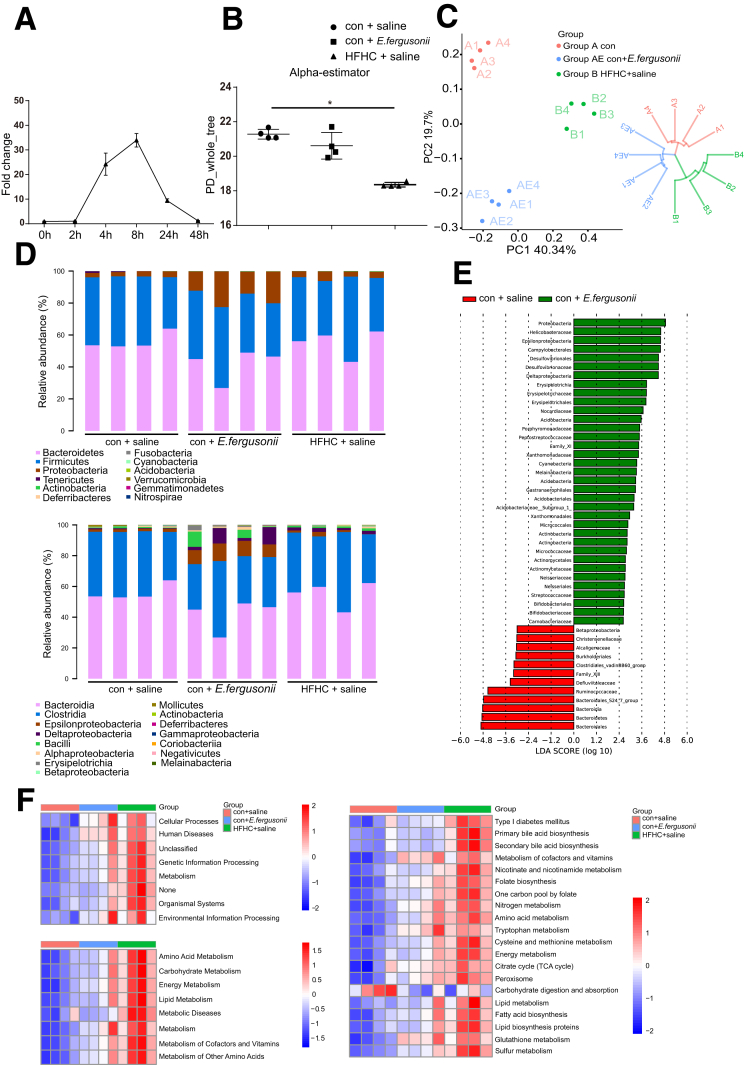
Figure 2.
E fergusonii altered the gut microbiome composition and functions. (A) E fergusonii was detected in feces from rats gavaged with E fergusonii (3∗109 colony-forming units), assessed by quantitative PCR. n = 6 per group. (B) Bacterial richness was estimated based on the Sobs estimator in rats from the con+saline, con+E fergusonii, and HFHC+saline groups. (C) β-diversity in rats from the different groups. (D) Relative abundance of bacteria at the phylum (upper) and class (lower) levels in rats from different groups. (E) Linear discriminant analysis effect size analysis between the con+saline and con+E fergusonii groups. (F) Phylogenetic Investigation of Communities by Reconstruction of Unobserved States analysis predicted gene functions in rats from different groups. ∗P < .05. LDA, linear discriminant analysis; PC, principal coordinate; PD, phylogenic diversity; TCA, tricarboxylic acid cycle.