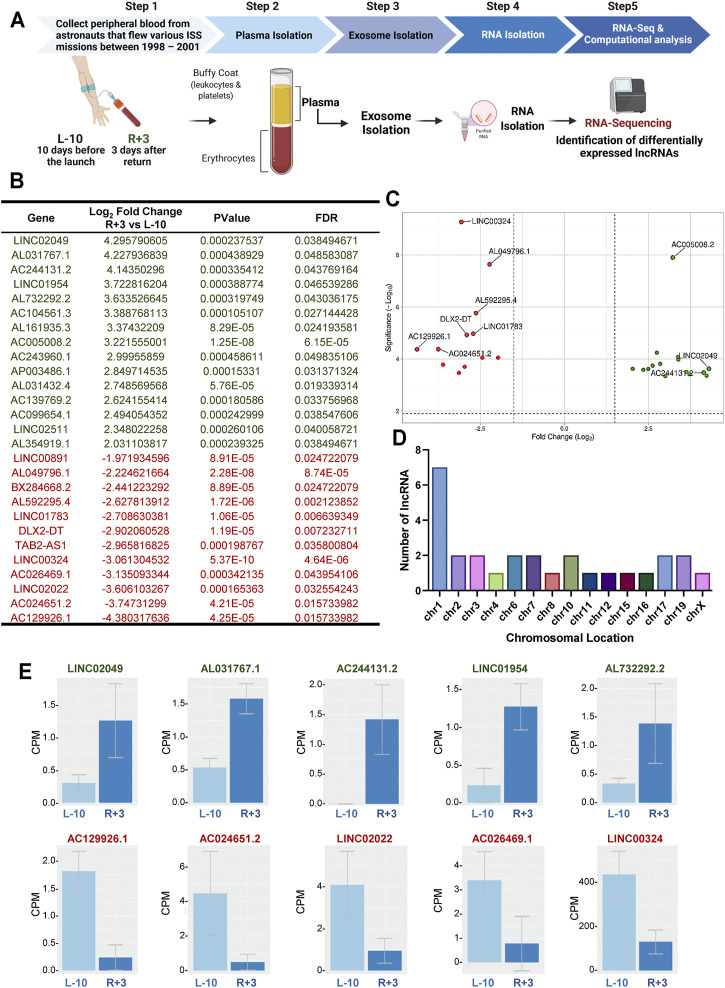
FIGURE 1.
Identification of 27 differentially regulated lncRNA in exosomes from astronauts 3-days post-landing. (A) Schematic representation of the experimental design. Blood was sampled at two different time points: 10 days before launch (L-10) and 3 days after return (R+3) from three different astronauts. Exosomes were isolated from blood plasma, and purified exosomal RNA was analyzed by RNA-Sequencing. (B) Computational analysis identified 27 differentially regulated lncRNA with a Log2 fold change >2, p < 0.001, and FDR <0.05, compared to baseline L-10. (C) Volcano plots showing Log2-fold changes for the 27 differentially regulated lncRNA and the statistical significance of each gene calculated after DEG analysis. Red points indicate significantly down-regulated genes; green points indicate up-regulated genes. (D) Chromosomal location of the lncRNA is shown. (E) Normalized counts, or the number of reads that align to a particular feature after correcting for sequencing depth and transcriptome composition bias, are shown for the top 5 up-regulated and down-regulated lncRNA in astronauts at L-10 and R+3.