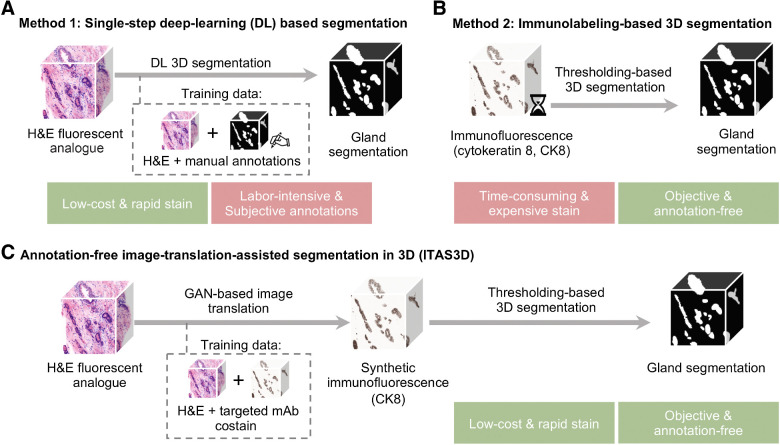
Figure 1.
General methods for 3D gland segmentation. A, A single-step DL segmentation model can be trained with imaging datasets of tissues labeled with a fluorescent analogue of H&E paired with manually annotated ground-truth segmentation masks. While H&E analogue staining is low-cost and rapid, manual annotations are labor-intensive (especially in 3D) and based on subjective human judgements. B, By immunolabeling a tissue microstructure with high specificity, 3D segmentations can be achieved with traditional CV methods without the need for manual annotations. While this is an objective segmentation method based on a chemical biomarker, immunolabeling large intact specimens is expensive and time-consuming due to the slow diffusion of antibodies in thick tissues. C, With ITAS3D, H&E-analogue datasets are computationally transformed in appearance to mimic immunofluorescence datasets, which enables the synthetically labeled tissue structures to be segmented with traditional CV methods. The image-sequence translation model is trained with a GAN based on paired H&E-analogue and immunofluorescence datasets. ITAS3D is rapid and low-cost (in terms of staining) as well as annotation-free and objective (i.e., biomarker-based).