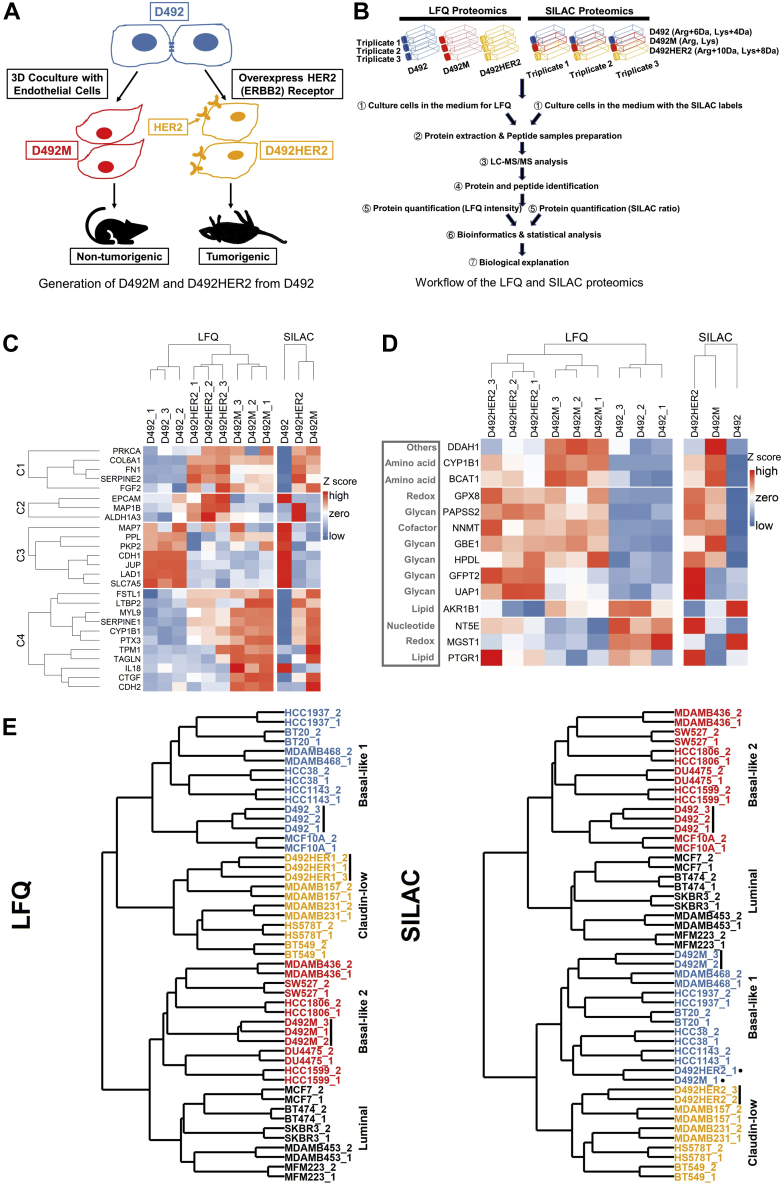
Fig. 1.
Overview of the study and the D492 EMT cell model.A, the process of generating D492M and D492HER2 from D492. The D492 cells were cocultured with endothelial cells (BRENCs or HUVECs) to generate spindle colonies that were subcultured to generate a new cell line D492M. The D492M cells are nontumorigenic. The HER2 (ERBB) receptor was overexpressed on the D492 cells to generate the D492HER2 cell line, and these cells can form tumors in mice. B, an overview of the whole proteomic experimental setup in this study from cell culture of D492, D492M, and D492HER2 to the bioinformatic and biological analysis of the LFQ and SILAC proteomic datasets. C, dysregulation of EMT markers in independent and published gene expression studies (GES) of EMT, which focused on different cell types and treatment modalities (42). D, dysregulation of EMT metabolic makers in the D492 cell model compared with the literature. There was a consistency between LFQ (left) and SILAC (right) except for NT5E. SILAC was consistent with the literature. HPDL, AKR1B1, and MGST1 were in an opposite trend compared with the literature. The mesenchymal metabolic signature (MMS) in the literature (44) was referred to in this analysis. For detailed descriptions of each EMT marker mentioned in Figure 1, C and D, please refer to the supplemental Data 4. E, classification of the D492 cell model. Using the iBAQ expression of proteins identified in both literature and this study, D492, D492M, and D492HER2 were clustered with other preclassified breast cell lines (45). LFQ (left) classified D492 as “Basal-like 1” (in blue), D492M as “Basal-like 2” (in red), and D492HER2 as “Mesenchymal-like/claudin-low” (in orange), while SILAC (right) classified D492 as “Basal-like 2” (in red), D492M as “Basal-like 1” (in blue), and D492HER2 also as “Mesenchymal-like/claudin-low” (in orange). The LFQ and SILAC raw data were quantified by the iBAQ quantification method in MaxQuant.