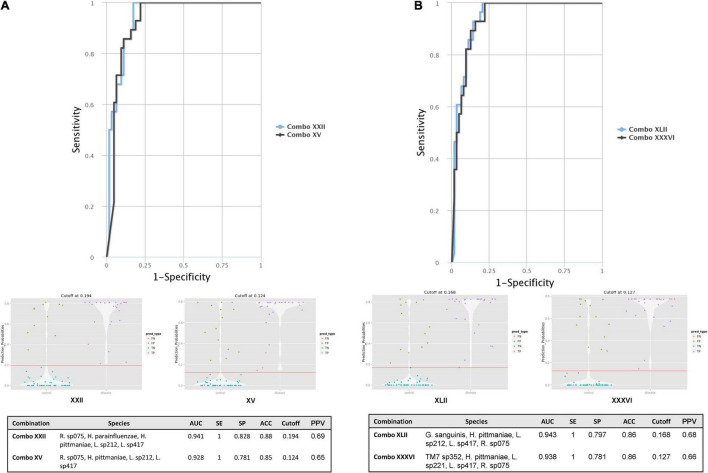
FIGURE 4.
HNC HPV+ vs. HNC HPV– CombiROC analysis using RA and log(RA + 1) transformed abundance data. (A) CombiROC analysis for biomarkers with AUC > 0.75 on RA data. (B) CombiROC analysis for biomarkers with AUC > 0.75 on log(RA + 1) data. Significant (α = 0.05) CombiROC program-generated receiver operator characteristic (ROC) curves for (A) RA data and (B) log(RA + 1) transformed abundance data of candidate microbial biomarkers (AUC > 0.75; n = 10 bacterial species) identified via MedCalc Software for Grp-All HPV+ vs. HPV– group are shown. (A) ROC curves, violin plots, and descriptive statistic summary table for the best two combinations from 10 potential markers using the RA are shown. “Combo XXII” consists of a combination of the markers Ruminococcaceae sp075, Haemophilus parainfluenzae, Haemophilis pittmaniae, Leptotrichia sp212, and Leptotrichia sp417 (ROC curve blue line). “Combo XV” contains markers Ruminococcaceae sp075, H. pittmaniae, Leptotrichia sp212, and Leptotrichia sp417 (ROC curve black line). (B) ROC curves, violin plots, and descriptive statistic summary table for the best two combinations from 10 potential markers using the log(RA + 1) transformed data are shown. “Combo XLII” contains potential markers Gemella sanguinis, H. pittmaniae, Leptotrichia sp212, and Ruminococcaceae sp075 (ROC curve blue line). “Combo XXXVI” consists of species probes TM7 G1 sp352, H. pittmaniae, Leptotrichia sp221, Leptotrichia sp417, and Ruminococcaceae sp075 (ROC curve black line). Cut-off refers to the optimal value or the highest true positive rate that has the lowest false positive rate. FN, false negative; FP, false positive; TN, true negative; TP, true positive; AUC, area under the curve; SE, sensitivity; SP, specificity; ACC, accuracy; PPV, positive predictive value.