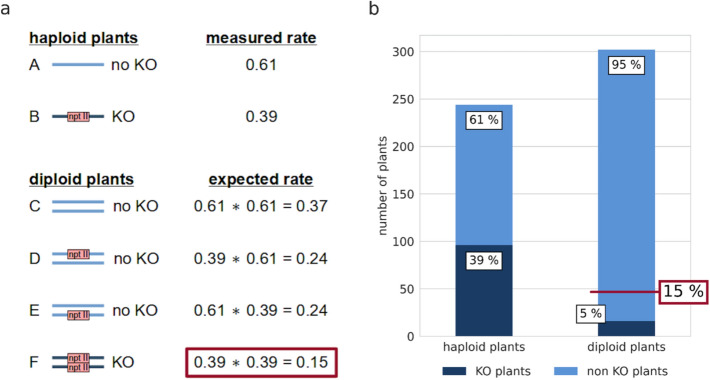
Fig. 5.
Diploid Physcomitrella cells show significantly lower GT rates. a: Estimation of the expected GT rate in diploid Physcomitrella plants computed from the measured GT rate in haploid plants. Shown are the observed rates of untransformed (A) and transformed (B) haploid plants as well as the expected rates of diploid plants having no integration of the cDNA construct in both chromosomes (C), having the cDNA construct integrated in only one chromosome (D and F) and having a full knock-out (KO) of the target locus in both chromosomes (F). The genomic loci are represented as solid lines and integration of a cDNA-construct containing the npt II cassette as selection marker is indicated. b: Comparison of GT rates in haploid and diploid Physcomitrella plants as determined via PCR analysis. For haploid plants, 96 out of 244 transformants are targeted KOs while for diploid plants, only 16 out of 302 transformants are targeted KOs on both chromosomes. The expected value of targeted KOs under the assumption of equal GT frequency for haploid and diploid plants is 15% (marked in red)