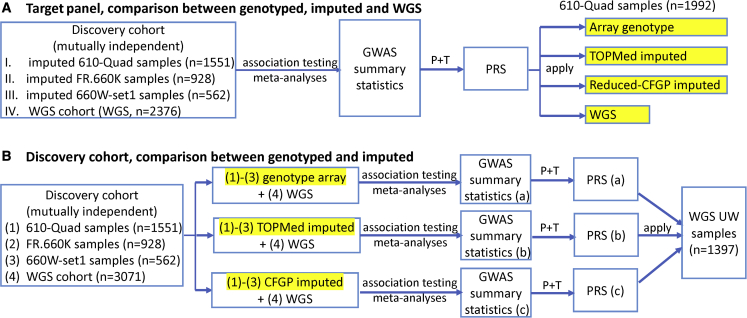
Figure 4.
Illustration of impact of imputation on PRS construction. (A) Imputation performed in target cohorts. We started with four independent discovery cohorts (I–III are TOPMed imputed data, IV is WGS data), performed association analysis for each subset separately and then meta-analyzed the association results. The meta-GWAS summary statistics was then used to construct PRS using the P+T method. The constructed PRS was applied to the same 1992 target samples but with four different marker densities (in yellow highlight): array genotype, TOPMed imputed, reduced CFGP imputed, or WGS data to compare the benefit of imputation in target cohort. (B) Imputation performed in discovery cohorts. We started with the same first three discovery cohorts as in A, but adopted three different marker sets (again in yellow highlight), as well as a fourth independent WGS cohort. We then performed association analysis and meta-analysis for each marker set, and constructed three different PRSs using the three different meta-GWAS summary statistics. The three PRSs were then applied to the same cohort to compare the performances.