Figure 3.
TLR2 interacts with the Wnt/β-catenin pathway and promotes the expression of splicing factors involved in the CD44 alternative splicing
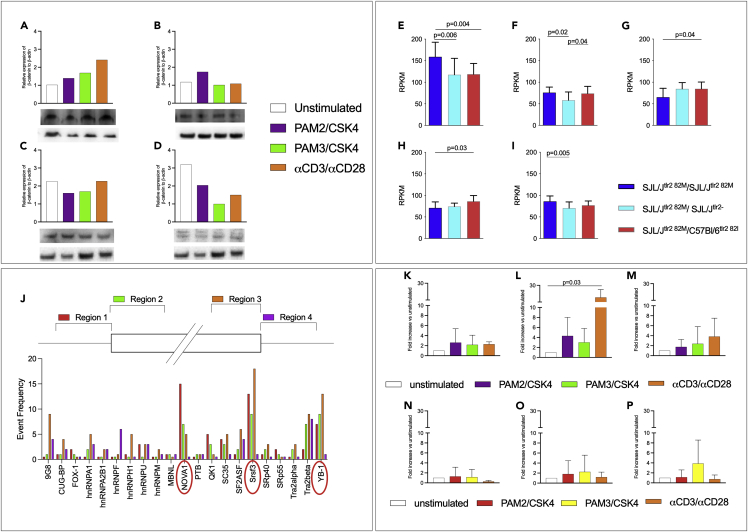
(A–D) Stimulation of TLR2/1 and TLR2/6 dimers blocks phosphorylation of β-catenin. CD4+CD62low T cells were enriched from the spleens of naive SJL/J mice (n = 3) and stimulated as described with PAM2/CSK4, PAM3/CSK4, or αCD3/αCD28 microparticles, for 1 (A and B) or 6 (C and D) hours. Proteins were purified and examined by Western blot for total (A and C) and phosphorylated (B and D) β-catenin (displayed data are the result of a single Western Blot assay).
(E–I) The wnt/β-catenin pathway is influenced by strain, TLR2 gene dose, and TLR2 haplotype. Five SJL/Jtlr2 82M/M (deep blue bars), six SJL/Jtlr2 82M/- (light blue bars), and six SJL/Jtlr2 82M/I (red bars) were challenged s.c. with CFA containing 50 μg/mouse of heat-inactivated Mtb. Microarray analysis was performed on CD4+ cells enriched from draining LN 4 days after immunization. Data are normalized versus the highest value obtained for each mRNA. Significance of the differences was evaluated by one-way ANOVA and Tukey HSD test. Values (mean + SD) are shown for Wnt10a (A), Wnt16 (B), Ctnnal1 (C), Wnt10b (D), and β-catenin (E).
(J) Splicing factors prediction through SFMAP software. Following the methodology of Paz et al. (Paz et al., 2010) and using the software SFMAP at http://sfmap.technion.ac.il, it has been possible to predict the splicing factors most probably involved in the alternative splicing of CD44v9-v10 and CD44v8-v10 (red circles indicate the most relevant ones).
(K–P) TLR2 stimulation changes mRNAs of the splicing factors NOVA-1, SRSF3, and YB-1 in SJL/J-derived but not in C57Bl/6-derived T cells. T cells isolated from SJL/J (K–M, n = 3) or C57Bl/6 (N–P, n = 4) were stimulated with PAM2/CSK4, PAM3/CSK4, or αCD3/αCD28 microparticles (data are shown as fold increase ± SD and normalized on unstimulated cells). As potentially involved in the alternative splicing of CD44 favoring the production of CD44v9-10 mRNA specific of three splicing factors, NOVA-1 (K andN), SRSF3 (L andO), and YB-1 (M andP) has been tested in RT-qPCR. Although differently depending on the stimulus, the three splicing factors resulted consistently upregulated in the SJL/J strain (one-way ANOVA statistical analysis showed a significant p = 0.03 for SRSF-3, unstimulated vs αCD3/αCD28 stimulated cells), while no relevant upregulation was observed in the C57Bl/6 strain, in which in most conditions is shown a downregulation of the same splicing factors.