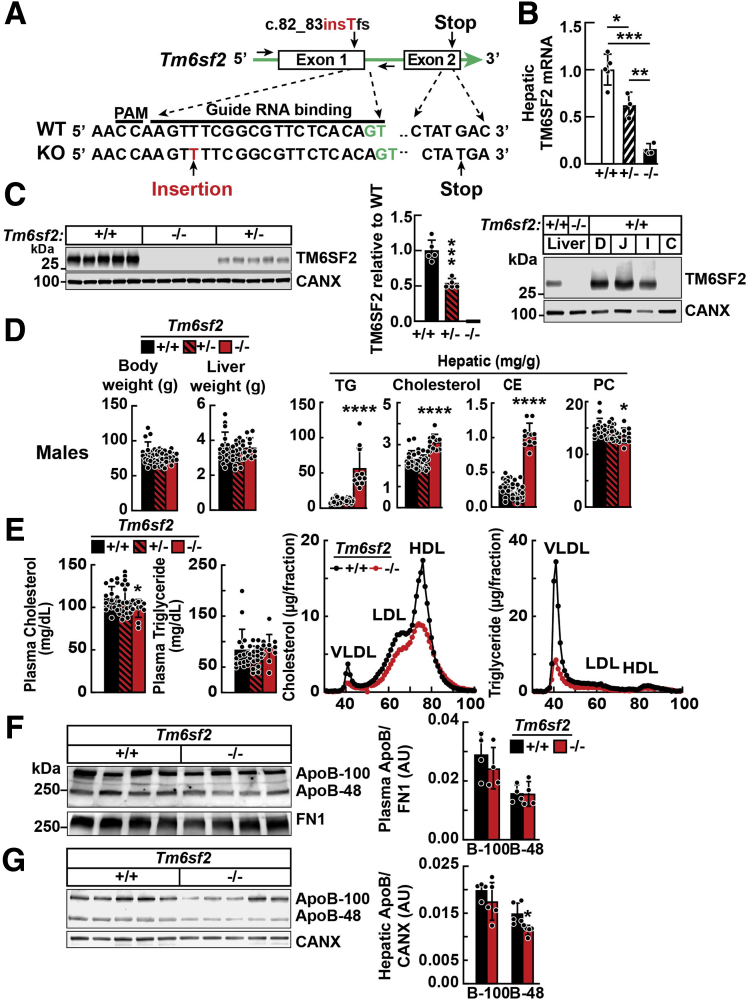
Figure 1.
Generation and characterization of Tm6sf2-/-rats using CRISPR/Cas9 technology. (A) Diagram of CRISPR/Cas9-targeted disruption of rat Tm6sf2 by insertion of T in exon 1 (c.80_81insT). Sequences of the guide RNA (gRNA) binding site, protospacer adjacent motif (PAM), and introns (green) are indicated. (B) TM6SF2 mRNA levels in female rat livers (n = 3–5/group, 3–4 wk) were determined using real-time PCR and normalized to levels of cyclophilin B mRNA. (C) Immunoblot of liver lysates (left) and enterocytes (right) from 4- to 5-week-old rats. (D) Body weights, liver weights, and hepatic lipids of chow-fed male rats (n = 11–19/group, 3–4 wk) after a 4-hour fast. (E) Plasma lipid levels were measured in the same rats as used in panel D (left). Plasma samples from WT and Tm6sf2-/- male rats (4/group, 5–6 wk) were pooled and size-fractionated by fast performance liquid chromatography. Cholesterol and TG levels were measured in each fraction (right). (F) Plasma (1 μL) was size-fractionated by 4%–15% gradient SDS–polyacrylamide gel electrophoresis. Levels of APOB were determined by immunoblot analysis using a rabbit polyclonal antibody (ab20737). Bars indicate means ± SD. Differences between groups were analyzed using the Student unpaired 2-tailed t test. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001, compared with the WT group. All experiments were repeated twice, and results were similar. C, colon; CANX, calnexin; D, duodenum; FN1, fibronectin; HDL, high-density lipoprotein; I, ileum; J, jejunum.