Abstract
DDX41 mutations are associated with hematologic malignancies including myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML), but the incidence in idiopathic cytopenia of undetermined significance (ICUS) is unknown. We investigated the incidence, genetic characteristics, and clinical features of DDX41 mutations in Korean patients with ICUS, MDS, or AML. We performed targeted deep sequencing of 61 genes including DDX41 in 457 patients with ICUS (n=75), MDS (n=210), or AML (n=172). Germline DDX41 mutations with causality were identified in 28 (6.1%) patients, of whom 27 (96.4%) had somatic mutations in the other position of DDX41. Germline origins of the DDX41 mutations were confirmed in all of the 11 patients in whom germline-based testing was performed. Of the germline DDX41 mutations, p.V152G (n=10) was most common, followed by p.Y259C (n=8), p.A500fs (n=6), and p.E7* (n=3). Compared with non-mutated patients, patients with a DDX41 mutation were more frequently male, older, had a normal karyotype, low leukocyte count, and hypocellular marrow at diagnosis. Three of the four ICUS patients with germline DDX41 mutations progressed to MDS. The incidence of DDX41 mutations in Korean patients was high and there was a distinct mutation pattern, in that p.V152G was a unique germline variant. ICUS harboring germline DDX41 mutations may be regarded as a hereditary myeloid neoplasm. Germline DDX41 mutations are not uncommon and should be explored when treating patients with myeloid malignancies.
Introduction
Inherited hematologic malignancies have been established in well-defined hereditary syndromes, which exhibit a particular phenotype, often present in childhood, or have a strong family history.1 There is also an increasing awareness of additional autosomal dominant genetic aberrations with predisposition to myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML), which are primarily sporadic diseases and typically present in older adults. The recently updated World Health Organization (WHO) classification defined myeloid neoplasms with germline predisposition2 and categorized familial myeloid neoplasms into three groups: those with an absence of pre-existing disorder or organ dysfunction (e.g., CEBPA or DDX41 mutations), those with a pre-existing platelet disorder (e.g., RUNX1 mutations), and those with dysfunction of other organs (e.g., GATA2 mutations). DDX41 mutations have recently joined the growing list of genetic alterations in familial myeloid malignancies.3,4 MDS or AML with germline DDX41 mutations usually occurs in the sixth decade of life or beyond, whereas most other cases of cancers with germline predisposing mutations typically develop in adolescence or early adulthood. 5-11
The DDX41 gene is located at 5q35.3 and encodes a DEAD-box RNA helicase, which is involved in pre-mRNA splicing, RNA processing, and ribosome biogenesis.12 Several mechanisms have been proposed to explain the contributions of DDX41 mutations to the development of hematologic malignancies. DDX41 mutations can: (i) cause aberrant mRNA splicing leading to exon retention or exon skipping, (ii) disrupt the STING-interferon pathway; and (iii) induce aberrant pre-rRNA trimming and ribosome biogenesis.3 DDX41 mutations include both germline and somatic mutations, with the latter being found in over half of the patients with germline mutations in the other allele of DDX41.5 In recent studies, germline DDX41 variants were found in 2.4% of 1,385 patients with MDS or AML,11 and germline or somatic DDX41 variants were found in 3.4% of 1,002 patients with myeloid neoplasms. 10
Following advances in genetic testing, clinical next-generation sequencing (NGS)-based leukemia panels are being increasingly used to identify somatic mutations to facilitate the diagnosis, improve prognostication, and select optimal treatment strategies in patients with hematologic malignancies. Some variants found in the panels can also be germline mutations in genes associated with hereditary hematopoietic malignancies.13-15 The myeloid leukemia panel used at our institute includes the DDX41 gene, and the frequencies of DDX41 mutations in Korean patients with MDS or AML appeared to be higher than the previously reported incidences. Importantly, DDX41 mutations have not been evaluated in patients with idiopathic cytopenia of undetermined significance (ICUS), which is a known precursor lesion of MDS. In this study, we investigated the incidence, genetic characteristics, and clinical features of DDX41 mutations in Korean patients with ICUS, MDS, or AML.
Methods
Patients
We included patients with ICUS, MDS, or AML whose bone marrow samples were collected between 2009 and 2019 at Asan Medical Center (Seoul, Korea). Patients with ICUS or lower-risk MDS were either prospectively enrolled (since January 2018) or retrospectively analyzed, while those with higher-risk MDS or AML were retrospectively analyzed. All patients in the study cohort were unrelated individuals, not including an index case and his or her family members. Diagnoses of MDS and AML were based on the WHO 2016 Classification.2 ICUS was defined by the proposed criteria of the 2007 Consensus Group:16 cytopenia in one or more of cell lineages for ≥6 months (hemoglobin <11 g/dL, neutrophils <1.5x109/L, and platelets <100x109/L) while excluding other causes of cytopenia such as a history of pelvic irradiation or cytotoxic chemotherapy, splenomegaly, heart failure or liver cirrhosis with portal hypertension, active viral infections, and a history of blood or bone marrow diseases. Clonal cytopenia of undetermined significance was defined as ICUS with myeloid neoplasm- related somatic mutations with a variant allele frequency ≥ 2%, or clonal karyotypic abnormalities. Myeloid neoplasm-related somatic mutations were based on those specified in the updated National Comprehensive Cancer Network guideline for MDS.17
The Institutional Review Board of Asan Medical Center approved the protocols of this study (2018-0042 and 2018-0048 [for prospective and retrospective analysis of patients with lowerrisk MDS or ICUS], 2019-0794 [for sequencing the DDX41 gene in DDX41-mutated patients and their family members], and 2020-0131 [for retrospective analysis of data from patients with higherrisk MDS or AML]), which was carried out in accordance with the 2008 Declaration of Helsinki.
Mutational and cytogenetic analysis
For rhe NGS assay, we prepared the sequencing libraries from genomic DNA using customized probes (Integrated DNA Technologies, Inc., Coralville, IA, USA) to capture and enrich the entire coding regions of 61 target genes (HEMEaccuTest DNA Target Enrichment kit; NGeneBio, Seoul, Korea) (Online Supplementary Table S1). We carried out sequencing on the MiSeqDx (Illumina, San Diego, CA, USA) with 2×150 bp, pairedend reads according to the manufacturer’s instructions. Initial read mapping was carried out against the human reference genome (hg19/GRCh37). We subsequently analyzed the sequencing data for variant calling using commercial software (CLC Genomics Workbench; QIAGEN Bioinformatics, Redwood City, CA, USA). We retained the potentially pathogenic variants by filtering out common polymorphisms (minor allele frequency in the population ≥1%) and sequencing/mapping errors, and by filtering in the known oncogenic variants based on the available population or cancer mutation-specific databases. We set the minimum cutoff of variant allele frequency at 2.0% for reporting. We performed the cytogenetic analysis using conventional G-banding techniques based on the analysis of 20 or more metaphase cells.
Germline variant confirmation and determination of causality
Variants with allele frequencies between 40% and 60% were considered to be probable germline mutations. We performed germline-based testing in 11 of 34 patients with probable germline DDX41 mutations using sorted blood T cells. This strategy of using sorted T cells was based on recent work confirming that T cells yield sufficient DNA and high rates of somatic variant calls in MDS. It was suggested that, given the challenge of obtaining skin biopsies, T cells would be preferential germline tissues for MDS genomic studies.18 Peripheral blood mononuclear cells were harvested by standard Ficoll (GE Healthcare, Sweden) density gradient centrifugation, and T cells were isolated using the Pan T Cell Isolation Kit, human (MACS Miltenyi Biotec, Auburn, CA, USA) according to the manufacturer’s instructions. The isolated T cells were analyzed with CD3-FITC using a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ, USA), and genomic DNA was purified by the QIAamp DNA mini kit (Qiagen, QIAGEN GmbH, Germany). The pathogenicity of probable germline DDX41 mutation was determined according to the guideline from the American College of Medical Genetics and Genomics (ACMG).19 The concurrence of a somatic DDX41 mutation was considered as strong evidence for causality. Thus, we classified germline DDX41 variants as “causal” if they were either pathogenic (or likely pathogenic) by the ACMG guideline or accompanied by a somatic DDX41 mutation regardless of the ACMG interpretation.
Statistical analysis
Categorical variables were compared using the χ2 test or Fisher exact test, and continuous variables were compared using the Mann-Whitney U-test or the Student t-test, as appropriate. Survival was calculated by the Kaplan-Meier method and the resulting survival curves were compared using the log-rank test (univariate analysis). The Kaplan-Meier survival curves were rendered as a graph using Prism version 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). In all analyses, the P-values were two-tailed and those <0.05 were considered statistically significant.
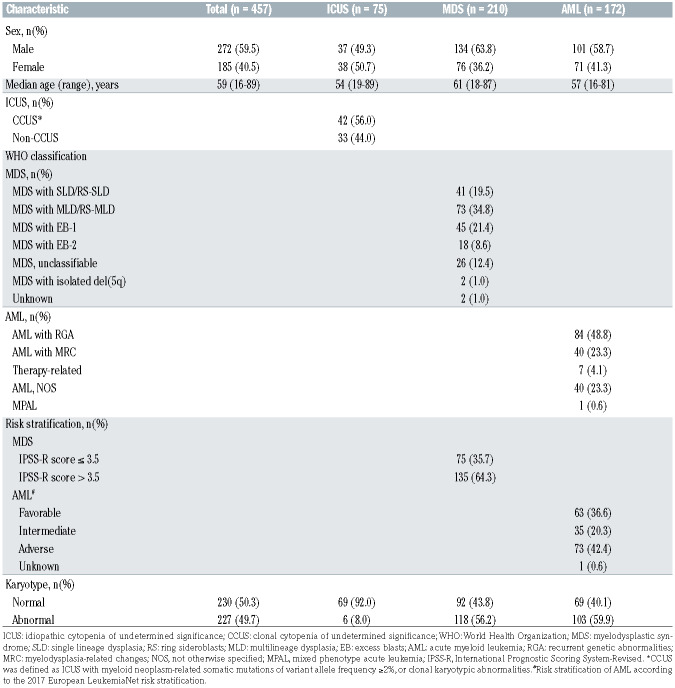
Table 1.
Patients’ characteristics at diagnosis.
Results
Patients’ characteristics
The clinical characteristics of the 457 included patients at diagnosis are shown in Table 1. There were 75 patients (16%) with ICUS, 210 (46%) with MDS and 172 (38% with AML. The median age at diagnosis was 59 years (range, 16-89), and 60% were men. Forty-two (56.0%) of the ICUS patients had clonal cytopenia of undetermined significance. Disease risk of the MDS patients was lowerrisk in 75 (35.7%) and higher-risk in 135 (64.3%) according to the Revised International Prognostic Scoring System (IPSS-R).20 Of the AML patients, 63 (36.6%), 35 (20.3%), and 73 (42.4%) were classified into favorable, intermediate, and adverse genetic risk categories, respectively, according to the 2017 European LeukemiaNet risk stratification. 21
Frequency and genetic characteristics of DDX41 mutations
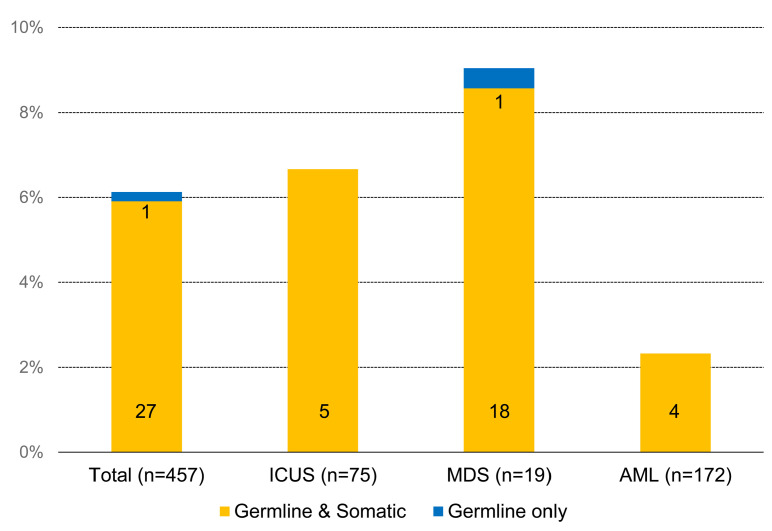
We detected genetic DDX41 mutations in 39 (8.5%) patients. Thirty-four (7.4%) patients had germline mutations, of whom 27 (79.4%) also had somatic mutations at the other position of DDX41. Five (1.1%) patients had somatic DDX41 mutations only. In 28 patients, the germline DDX41 mutations were considered causal and only these patients were included in further analyses. The frequency of the causal germline DDX41 mutations was 6.1% (28 of 457); 6.7% (5 of 75) in ICUS, 9.0% (19 of 210) in MDS, and 2.3% (4 of 172) in AML (Figure 1). Detailed information on the DDX41 variants, concurrent mutations of other genes, and karyotypes in the 28 patients are provided in Online Supplementary Table S2. Germline origins of the DDX41 mutations were confirmed in all of the 11 patients who underwent germline-based testing (p.V152G in 5, p.Y259C in 3, p.A500fs in 2, and p.L328R in 1).
Figure 1.
Frequency of DDX41 mutations according to the type of hematologic malignancy. ICUS: idiopathic cytopenia of undetermined significance; MDS: myelodysplastic syndrome; AML: acute myeloid leukemia.
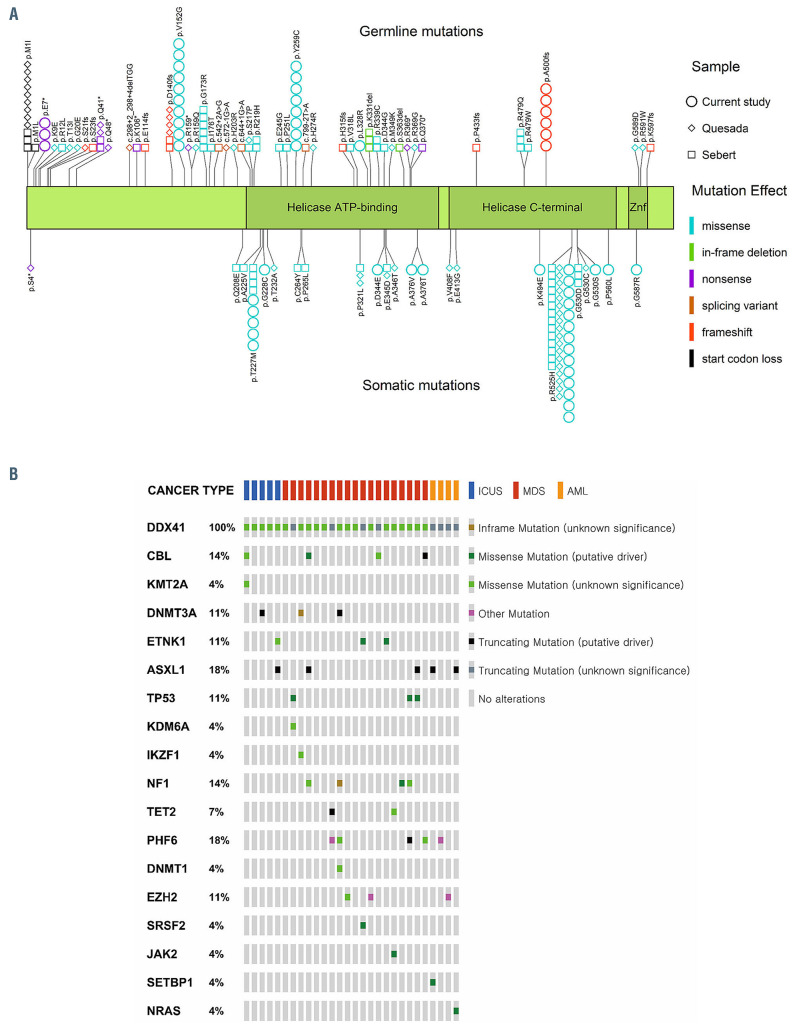
Of the 55 DDX41 mutations detected in this study, 28 were germline and the other 27 appeared to be somatic. All of the somatic mutations were missense, whereas germline mutations were missense in 19 (67.9%) cases, frameshift in six (21.4%), and nonsense in three (10.7%). The majority of somatic mutations were located in the helicase C or C-terminal domain (n=18, 66.7%), whereas the majority of germline mutations were in the helicase ATP-binding or N-terminal domain (n=22, 78.6%; P=0.001) (Figure 2A). Of the germline DDX41 mutations, p.V152G (n=10, 35.7%) was the most common, followed by p.Y259C (n=8, 28.6%), p.A500fs (n=6, 21.4%), p.E7 *(n=3, 10.7%), and p.L328R (n=1, 3.6%).
Two germline variants (p.A500fs and p.E7*) were classified as pathogenic according to the ACMG guideline. The other germline variants (p.V152G, p.Y259C, and p.L328R) were classified as being of uncertain significance, but were considered causal when accompanied by somatic DDX41 mutations (Online Supplementary Table S3). Notably, four mutations (p.V152G, p.Y259C, p.A500fs and p.E7*) were found at a significantly higher frequency in the study patients than in healthy Koreans, as shown by high odds ratios (38.5, 17.3, 49.6 and 26.5, respectively) (Online Supplementary Table S4). Two germline mutations (p.V152G and p.Y259C) were only detected in ICUS/MDS (75.0%) and not in AML (0%), whereas p.A500fs and p.E7* were detected in both ICUS/MDS and AML groups (Online Supplementary Table S5). Of the 27 somatic DDX41 mutations, p.R525H (n=14, 51.9%) was the most common, followed by p.T227M (n=5, 18.5%), and the remaining eight somatic DDX41 mutations were detected in one (3.7%) patient each. The somatic p.R525H variant was less frequently associated with the germline p.V152G variant (3 of 10) than with p.Y259C (6 of 8) or p.A500fs (4 of 6), whereas the somatic p.T227M variant tended to be more frequently associated with p.V152G (4 of 10) than with p.Y259C (1 of 8) or p.A500fs (0 of 6) (Online Supplementary Table S6).
Twenty-two (78.6%) of the 28 patients with mutations in DDX41 had concurrent mutations in other genes. Genes mutated in over 10% of the patients were PHF6 and ASXL1 (5 patients [17.9%] each), followed by CBL and NF1 (4 patients [14.3%] each), and DNMT3A and TP53 (3 patients [10.7%] each) (Figure 2B; Online Supplementary Table S2). We observed six variants of the PHF6 gene in five patients with DDX41 germline mutations: p.M1T and p.R116* in one patient, and p.G248D, p.C20F, p.M1T and p.M1V in one patient each. Interestingly, PHF6 p.M1T/V variants were detected in only three patients harboring DDX41 germline mutations among the whole study population of 457 patients.
Clinical features and outcomes of the patients with DDX41 mutations
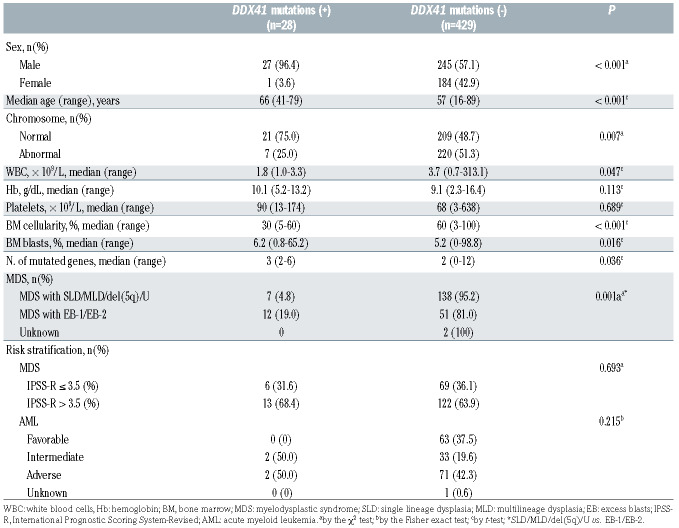
There was a male predominance among the DDX41-mutated patients (96.4% vs. 57.1%; P<0.001), and the patients with this mutation tended to be older (median 66 vs. 57 years; P<0.001), and were more likely to have a normal karyotype (75.0% vs. 48.7%; P=0.007), lower white blood cell count (median 1.8 vs. 3.7×109/L; P=0.047), and lower marrow cellularity (median 30% vs. 60%; P<0.001) at diagnosis compared with the non-mutated patients (Table 2). Among patients with MDS, the DDX41 mutations were significantly more frequent in the MDS subtypes with excess blasts (EB)-1 and EB-2, compared to other categories with bone marrow blasts <5%, although the mutation frequencies were not significantly different between patients with lower risk or higher risk according to the IPSS-R (Table 2). Of 23 MDS or AML patients with causal germline DDX41 mutations, data regarding blood counts before diagnosis were available for 16 patients, and all 16 patients had a history of cytopenia at least 1 year prior to diagnosis.
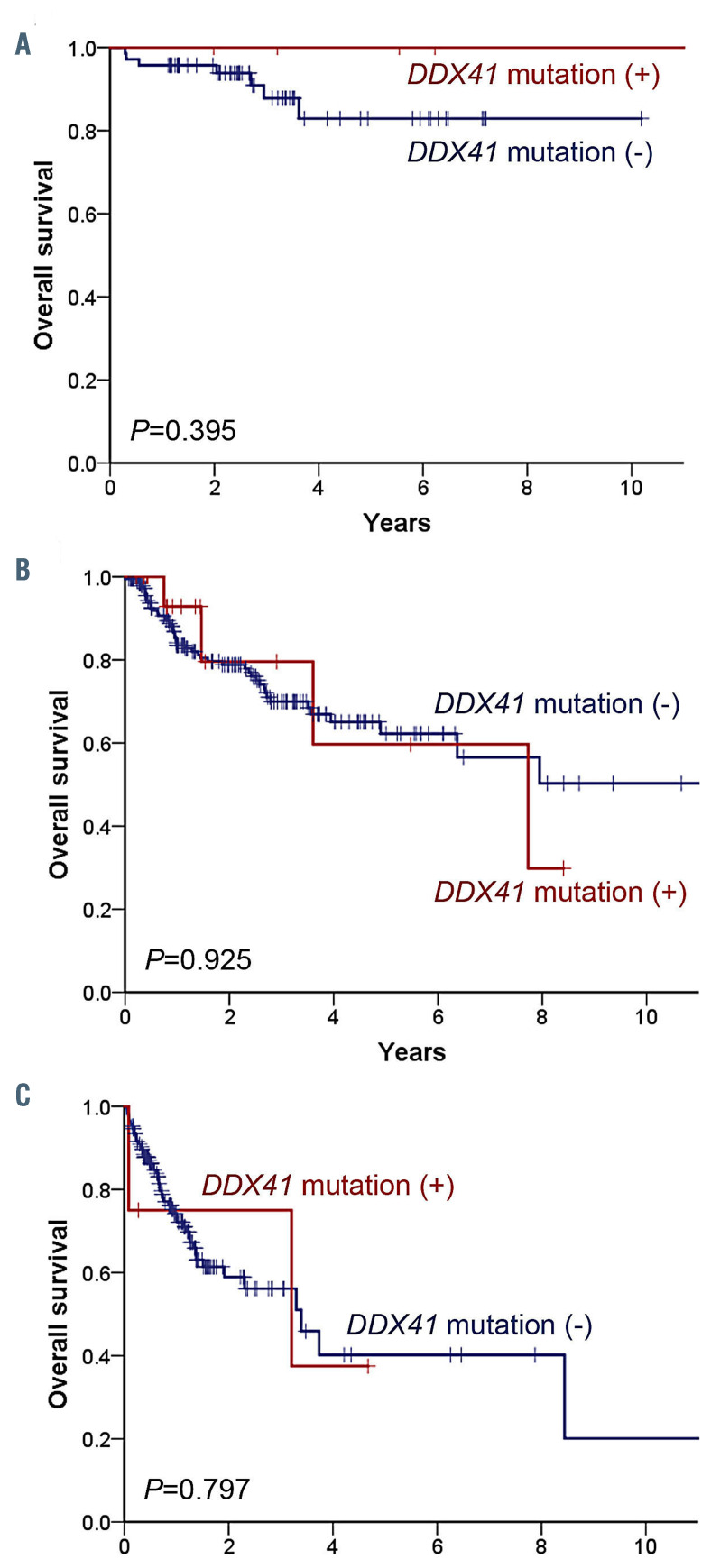
During the median follow-up of 25.5 months, 116 patients (7 ICUS, 55 MDS, and 54 AML) died. The 5-year overall survival rate was 60.8% in the overall population and 84.6%, 62.2%, and 38.9% in patients with ICUS, MDS, and AML, respectively. There was no significant correlation between overall survival and the presence of DDX41 mutations in each disease category of ICUS, MDS, and AML (Figure 3) as well as in the total study population (Online Supplementary Figure S1).
Table 2.
Comparison of clinical features according to the presence of germline DDX41 mutations.
Online Supplementary Table S7 shows the clinical course of each patient with a DDX41 mutation. Clinical courses could be followed up in four of the five ICUS patients with probable germline DDX41 mutations, and notably, three of these four patients showed disease progression to MDS EB-1 (n=2; 77.9 and 17.6 months after ICUS diagnosis) or MDS EB-2 with a gain of PTPN11 mutation (n=1; 9 months after ICUS diagnosis) during the follow-up. Another ICUS patient with a germline DDX41 mutation had a son with Hodgkin lymphoma.
Discussion
In our cohort of 457 patients with ICUS, MDS, or AML, 6.1% of the patients carried causal germline DDX41 mutations, which is a higher incidence than those found in previous studies which ranged between 0.8% and 3.9% in patients with myeloid malignancies (mostly MDS and AML).5,6,10,11,22 In a study comparing the clinical and genetic characteristics of DDX41 mutations in AML and MDS patients between two ethnically distinct populations, germline DDX41 mutations were found in 3.9% of a Japanese cohort and in 0.8% of a Caucasian cohort.22 Therefore, there seems to be an ethnic difference in the incidence of DDX41 mutations in patients with myeloid neoplasms between Asian and Western patients. In contrast, the clinical features of our DDX41 -mutated patients, such as male predominance, old age at presentation, 5,6,10,11,23 hypocellular marrow,3,4,6 leukopenia,6 and frequent normal cytogenetics3-6,11 were similar to those reported in other ethnic populations. The DDX41 mutations did not show significant associations with survival outcomes.
There are several noteworthy findings in our study regarding the genetic characteristics of DDX41 mutations. First, the germline mutations were mostly N-terminal variants (78.6%), whereas somatic mutations were mostly C-terminal variants (66.7%). This finding is consistent with the observations in two recent studies.10,11 The N-terminal region of DDX41 has the helicase ATP binding domain,24,25 and the structural rearrangement in the N-terminal region may change the conformation of the ATP-binding site and eventually decrease ATP-binding ability.24 In contrast, the helicase C-terminal domain is involved in ATP hydrolysis.24,25 Therefore, genetic alterations in the N-terminal region may cause greater susceptibility to protein hypofunction than those in the C-terminal region. Second, the patterns of germline DDX41 mutations in our Korean population were distinct from those in Western populations10,11 or even other Asian populations. 22,23 The germline DDX41 mutations (p.V152G, p.Y259C, p.A500fs, p.E7*, and p.L328R) in our study are totally different from those reported in Western populations (p.M1I, p.D140fs, p.G173R, and Q41*). Korean and Japanese patients shared three major germline DDX41 variants (p.Y259C, p.A500fs, p.E7*),22 but p.V152G was only found in Koreans and not in Japanese or other ethnic populations. Third, we observed the exclusive presence of PHF6 p.M1T/V variants in three patients with probable germline DDX41 mutations. These variants potentially cause a complete lack of protein production as a consequence of start codon loss and are causative germline mutations of the Börjeson-Forssman-Lehmann syndrome.26-28 Thus, our findings suggest that the same genetic mutation can induce both hereditary diseases and sporadic cancer, as exemplified by mutations in ETV6.29 The possible association between PHF6 p.M1T/V variants and germline DDX41 mutations should be investigated further.
Figure 2.
Distribution of DDX41 mutations and concurrent mutations in other genes. (A) Distribution of DDX41 mutations detected in the current study and two previous studies (Quesada et al.10 and Sebert et al.11). This figure shows the differences in positional distribution (N-terminal skewed vs. C-terminal skewed) and mutational effects (variable vs. missense-dominated) between germline and somatic mutations. The protein structure of DDX41 was based on the RefSeq accession number of NM_016222.3 and the UniProtKB entry of Q9UJV9: the 622 amino acid long protein comprises the helicase ATP-binding domain (position 212-396), the helicase C-terminal domain (position 407-567), and a zinc finger domain (position 580-597). Different colors indicate different effects of mutations: light blue, missense mutation; light green, inframe indel; purple, nonsense mutation; brown, splicing mutation; red, frameshift mutation; black, start codon loss. Different shapes represent the three studies: square, Sebert et al.11 diamond, Quesada et al.10 circle, current study. (B) Concurrent mutations of other genes identified in bone marrow samples from DDX41-mutated patients. The types of genetic alterations and diseases are presented in the legend.
Germline mutations that predispose an individual to MDS or AML may also contribute to the development of ICUS, but the genetic predisposition to ICUS has not been systematically investigated. In a recent study of germline DDX41 mutations in adult patients with MDS or AML, 45.5% of patients with pathogenic germline DDX41 mutations had a previous history of cytopenia before the diagnosis of MDS or AML, and the preexisting cytopenia might indicate the presence of ICUS in these patients.11 We also observed similar findings. Furthermore, five (6.7%) of 75 ICUS patients had causal germline DDX41 mutations, three of whom progressed to MDS. Our study shows that germline DDX41 mutations are not uncommon in ICUS patients. Our findings do not indicate that the germline DDX41 mutations contribute to the progression of ICUS to MDS, but instead do suggest that ICUS patients harboring such variants may be considered as having a hereditary myeloid neoplasm.
Our observations highlight the potential oncogenic role of germline DDX41 mutations in the pathogenesis of ICUS/MDS in comparison with AML. First, the patients with ICUS/MDS carried germline DDX41 mutations more frequently than did AML patients and germline missense mutations were highly enriched in ICUS/MDS rather than in AML. These findings might support the notion that less disruptive variants are associated with a milder phenotype in the disease spectrum. Second, only one (3.6%) of the 28 patients with germline DDX41 mutations carried a mutation in the splicing factor gene. This finding is in line with previous observations that splicing factor gene mutations were largely mutually exclusive with DDX41 mutations. 5,10 DDX41 interacts with core splicing proteins such as SF3B, U2 complex, PRPF8 scaffold protein, and U5 complex, indicating that spliceosomal proteins are the top functional group associated with DDX41.5,25 Genetic alterations of the splicing components affect the 3’-splice site recognition during pre-mRNA processing and are involved in the pathogenesis of myelodysplasia.30 This indicates that mutant DDX41 can have an oncogenic role in MDS via aberrant mRNA splicing with the assumption that mutations in these splicing factors have an impact on the pathways of downstream oncogenes and tumor suppressor genes.3
Donor-derived leukemia has been reported in several families with germline DDX41 mutations; in all such cases, donors had the same type of germline DDX41 variants as the respective recipients.31,32 In our study, DDX41 mutations (germline p.E7* and somatic p.G228C) were found in a 60-year-old man with high-risk MDS (#12). No HLA-matched sibling or unrelated donor was available, and his two adult offspring had the same DDX41 mutation (p.E7*). Fortunately, the patient’s HLA-haploidentical brother did not carry the DDX41 mutation, and the patient could undergo haploidentical hematopoietic stem cell transplantation from him. Considering that the risk of malignancy in DDX41 carriers is yet to be determined, we calculated the odds ratios of major germline DDX41 variants detected in our study (Online Supplementary Table S4). Nevertheless, an extensive population-based study is needed to obtain more reliable data that may be useful in establishing genetic counseling guidelines for germline DDX41 variants, which are currently available only for donor selection in allogeneic hematopoietic stem cell transplantation.
Figure 3.
Overall survival of patients with different hematologic disorders according to DDX41 mutation status. (A-C) Overall survival of patients with idiopathic cytopenia of undetermined significance (A), myelodysplastic syndrome (B) or acute myeloid leukemia (C) according to whether they had DDX41 mutations (red) or not (blue).
NGS-based targeted genotyping for somatic mutations can identify patients who are at risk of hereditary hematopoietic malignancies. In a recent study, of 25 pathogenic or likely pathogenic variants with variant allele frequency >40% in 24 patients with germline tissues available, six variants (24%) were of germline origin – three DDX41 variants, two GATA2 variants, and one TP53 variant; DDX41 had a 100% diagnostic yield for pathogenic germline variants in that study.33 In another study, targeted NGS showed that 17 patients had putative germline DDX41 variants with a variant allele frequency >40%, all of which were of germline origin.11 We were also able to confirm germline origin in all of the 11 patients with probable germline DDX41 mutations. In cases in which germline samples are not available, NGSbased leukemia panels seem to predict germline DDX41 variants with high probability. However, it is worth mentioning that NGS-based panels may fail to detect deletions or gene rearrangements that are responsible for the predisposition syndrome.
Our study has some limitations. The number of patients included in the study was relatively small, and this might have had an impact on the analysis for clinical associations of DDX41 mutations with clinical outcomes. Family history was not systematically collected in this study, although such information is helpful in pinpointing the pathogenicity of sequence variants. We did not perform functional studies to demonstrate that sequence variants detected in this study had a deleterious effect in vivo. Experimental data can be useful to support pathogenicity, particularly for missense variants of uncertain significance. We acknowledge that these limitations may hamper the precise variant classification based on the ACMG guideline. However, the concurrence of germline and somatic DDX41 mutations was a recurrent finding across recent studies.5,6,10,11 This also has provided another illustration that germline alterations predispose to the acquisition of somatic mutations in the same genes which act as a second hit being associated with cancer development as demonstrated by JAK2, CEBPA, and RUNX1 mutations. 34,35 Therefore, in cases harboring a germline DDX41 mutation, the acquisition of a somatic DDX41 mutation should be considered as strong evidence for causality. Lastly, our data may not reflect the whole Korean population, although study patients included in this study come from all across the Korean peninsula.
In conclusion, our results delineate the unique ethnic features of DDX41 mutations in Korean patients, such as higher incidence and different patterns, compared with patients from Western countries or other Asian countries. Specifically, the most common germline mutation in our cohort was p.V152G, which was not found in previous studies in other ethnicities. Our results suggest that ICUS harboring germline DDX41 mutations may be regarded as a hereditary myeloid neoplasm. Germline DDX41 mutations may be predicted with a high probability by using clinical NGS-based leukemia panels based on variant allele frequency levels and public databases. Germline DDX41 mutations are not uncommon and should be explored when treating patients with myeloid malignancies.
Supplementary Material
Acknowledgments
We thank Dr. Joon Seo Lim from the Scientific Publications Team at Asan Medical Center for his editorial assistance in preparing this manuscript.
Funding Statement
Funding: This research was supported by a Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (NRF-2017R1E1A1A01074383). The biospecimens and data used in this study were provided by Asan Bio-Resource Center, Korea Biobank Network (2018-08).
References
- 1.Rafei H, DiNardo CD. Hereditary myeloid malignancies. Best Pract Res Clin Haematol. 2019;32(2):163-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016; 127(20):2391-2405. [DOI] [PubMed] [Google Scholar]
- 3.Cheah JJC, Hahn CN, Hiwase DK, Scott HS, Brown AL. Myeloid neoplasms with germline DDX41 mutation. Int J Hematol. 2017;106(2):163-174. [DOI] [PubMed] [Google Scholar]
- 4.Maciejewski JP, Padgett RA, Brown AL, Muller-Tidow C. DDX41-related myeloid neoplasia. Semin Hematol. 2017;54(2):94-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polprasert C, Schulze I, Sekeres MA, et al. Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell. 2015; 27(5):658-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewinsohn M, Brown AL, Weinel LM, et al. Novel germ line DDX41 mutations define families with a lower age of MDS/AML onset and lymphoid malignancies. Blood. 2016;127(8):1017-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardoso SR, Ryan G, Walne AJ, et al. Germline heterozygous DDX41 variants in a subset of familial myelodysplasia and acute myeloid leukemia. Leukemia. 2016; 30(10):2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li R, Sobreira N, Witmer PD, Pratz KW, Braunstein EM. Two novel germline DDX41 mutations in a family with inherited myelodysplasia/acute myeloid leukemia. Haematologica. 2016;101(6): e228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vairo FPE, Ferrer A, Cathcart-Rake E, et al. Novel germline missense DDX41 variant in a patient with an adult-onset myeloid neoplasm with excess blasts without dysplasia. Leuk Lymphoma. 2019;60(5):1337-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quesada AE, Routbort MJ, DiNardo CD, et al. DDX41 mutations in myeloid neoplasms are associated with male gender, TP53 mutations and high-risk disease. Am J Hematol. 2019;94(7):757-766. [DOI] [PubMed] [Google Scholar]
- 11.Sébert M, Passet M, Raimbault A, et al. Germline DDX41 mutations define a significant entity within adult MDS/AML patients. Blood. 2019;134(17):1441-1444. [DOI] [PubMed] [Google Scholar]
- 12.Linder P. Dead-box proteins: a family affair- -active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34(15):4168-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiNardo CD, Routbort MJ, Bannon SA, et al. Improving the detection of patients with inherited predispositions to hematologic malignancies using next-generation sequencing-based leukemia prognostication panels. Cancer. 2018;124(13):2704-2713. [DOI] [PubMed] [Google Scholar]
- 14.Drazer MW, Kadri S, Sukhanova M, et al. Prognostic tumor sequencing panels frequently identify germ line variants associated with hereditary hematopoietic malignancies. Blood Adv. 2018;2(2):146-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guidugli L, Johnson AK, Alkorta-Aranburu G, et al. Clinical utility of gene panel-based testing for hereditary myelodysplastic syndrome/ acute leukemia predisposition syndromes. Leukemia. 2017;31(5):1226-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valent P, Horny H-P, Bennett JM, et al. Definitions and standards in the diagnosis and treatment of the myelodysplastic syndromes: consensus statements and report from a working conference. Leuk Res. 2007;31(6):727-736. [DOI] [PubMed] [Google Scholar]
- 17.NCC Network. Myelodysplastic syndrome (Version 2.2020). https://www.nccn.org/professionals/physician_gls/PDF/mds.pdf Accessed May 10, 2020. [Google Scholar]
- 18.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Padron E, Ball MC, Teer JK, et al. Germ line tissues for optimal detection of somatic variants in myelodysplastic syndromes. Blood. 2018;131(21):2402-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenberg PL, Tuechler H, Schanz J, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120(12):2454-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeda J, Yoshida K, Makishima H, et al. Genetic predispositions to sporadic myeloid neoplasms caused by germline DDX41 mutations in Asian and Caucasian populations. Haematologica. 2016; 101 (s1):66-67. [Google Scholar]
- 23.Polprasert C, Takeda J, Niparuck P, et al. Novel DDX41 variants in Thai patients with myeloid neoplasms. Int J Hematol. 2020;111(2):241-246. [DOI] [PubMed] [Google Scholar]
- 24.Omura H, Oikawa D, Nakane T, et al. Structural and functional analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide. Sci Rep. 2016;6(1):1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang Y, Zhu Y, Liu Z-J, Ouyang S. The emerging roles of the DDX41 protein in immunity and diseases. Protein Cell. 2017; 8(2):83-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crawford J, Lower KM, Hennekam RC, et al. Mutation screening in Börjeson- Forssman-Lehmann syndrome: identification of a novel de novo PHF6 mutation in a female patient. J Med Genet. 2006; 43(3):238-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst A, Le VQ Højland AT, et al. The PHF6 mutation c. 1A> G; PM1V causes Börjeson- Forsman-Lehmann syndrome in a family with four affected young boys. Mol Syndromol. 2015;6(4):181-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Todd MA, Ivanochko D, Picketts DJ. PHF6 degrees of separation: the multifaceted roles of a chromatin adaptor protein. Genes. 2015;6(2):325-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feurstein S, Godley LA. Germline ETV6 mutations and predisposition to hematological malignancies. Int J Hematol. 2017; 106(2):189-195. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64-69. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi S, Kobayashi A, Osawa Y, et al. Donor cell leukemia arising from preleukemic clones with a novel germline DDX41 mutation after allogenic hematopoietic stem cell transplantation. Leukemia. 2017;31(4):1020. [DOI] [PubMed] [Google Scholar]
- 32.Berger G, van den Berg E, Sikkema-Raddatz B, et al. Re-emergence of acute myeloid leukemia in donor cells following allogeneic transplantation in a family with a germline DDX41 mutation. Leukemia. 2017;31(2): 520. [DOI] [PubMed] [Google Scholar]
- 33.Drazer MW, Kadri S, Sukhanova M, et al. Prognostic tumor sequencing panels frequently identify germ line variants associated with hereditary hematopoietic malignancies. Blood Adv. 2018;2(2):146-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kilpivaara O, Mukherjee S, Schram AM, et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2 V617F-positive myeloproliferative neoplasms. Nat Genet. 2009;41(4):455-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown AL, Hahn CN, Scott HS. Secondary leukemia in patients with germline transcription factor mutations (RUNX1, GATA2, CEBPA). Blood. 2020;136(1):24-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.