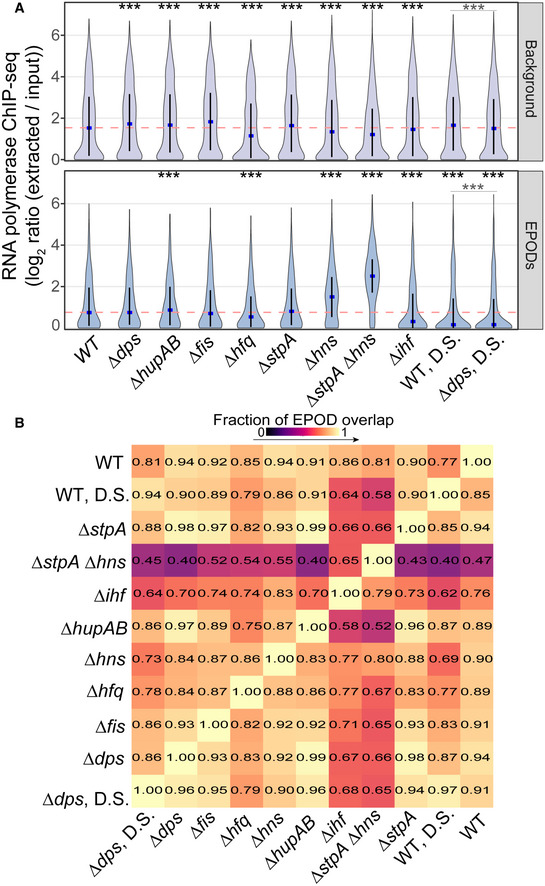
Average RNA polymerase occupancy was calculated across intergenic regions within WT EPODs and background. Similar to Fig
2D, the blue dots denote the median and the black line displays the interquartile ranges in each condition. The dashed pink line represents the WT median. (*) indicate the Wilcoxon rank‐sum
P value comparing the change in median versus WT for each condition that has been adjusted using the Benjamini and Hochberg method (against a null hypothesis of no difference in pseudomedians). The gray line denotes the same comparison between the D.S. conditions.
P value < 0.0005 = ***. Data were averaged across 2 (
hupAB,
fis,
stpA,
ihf,
dps), 3 (WT,
hfq, WT DS,
dps DS),or 4 (
hns,
stpA/hnS) biological replicates.