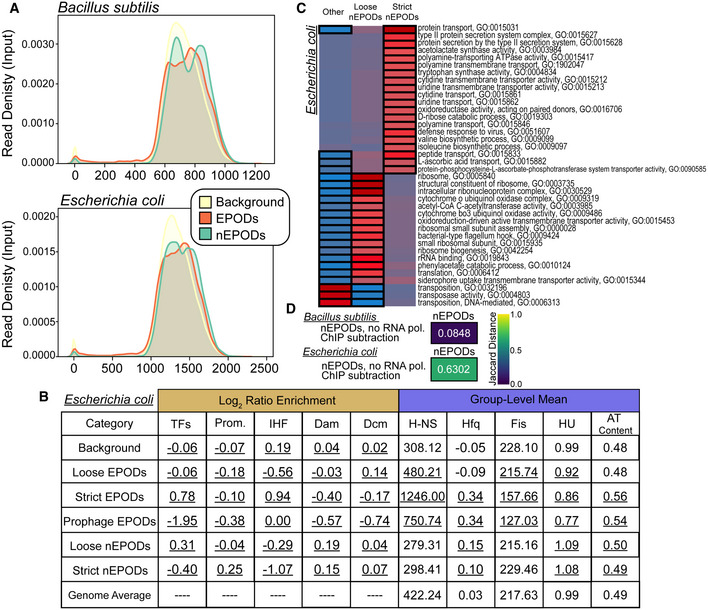
Kernel density plots displaying the input read density Bacillus subtilis and Escherichia coli in regions called as EPODs (orange), nEPODs (green), and Background (yellow).
Assessment of Escherichia coli nEPODs compared to other regions. Log2 ratio enrichment: The ratio of the number of TFs, promoters, or motifs in a given category to the total number of TFs, promoters, or motifs. A chi‐squared test was performed, and all categories were significantly associated with each class; values underlined had a P‐value < 0.05. Group‐level mean: The 500‐bp rolling mean for the binding of each NAP was used to calculate the group‐level means for across each category and compared with the overall average for the genome. Permutation based P‐values were calculated comparing each class versus the background. The values underlined had a P‐value < 0.05.
Escherichia coli nEPODs overlap highly transcribed gene categories.
Jaccard distance comparing nEPODs called with and without RNA polymerase ChIP‐seq subtraction. The high Jaccard distance in Escherichia coli indicates that nEPODs change dramatically with and without the inclusion RNA polymerase binding, and therefore RNA polymerase associated. However, locations of nEPODs in Bacillus subtilis are unchanged with RNA polymerase binding and are distinct, protein occupied regions.