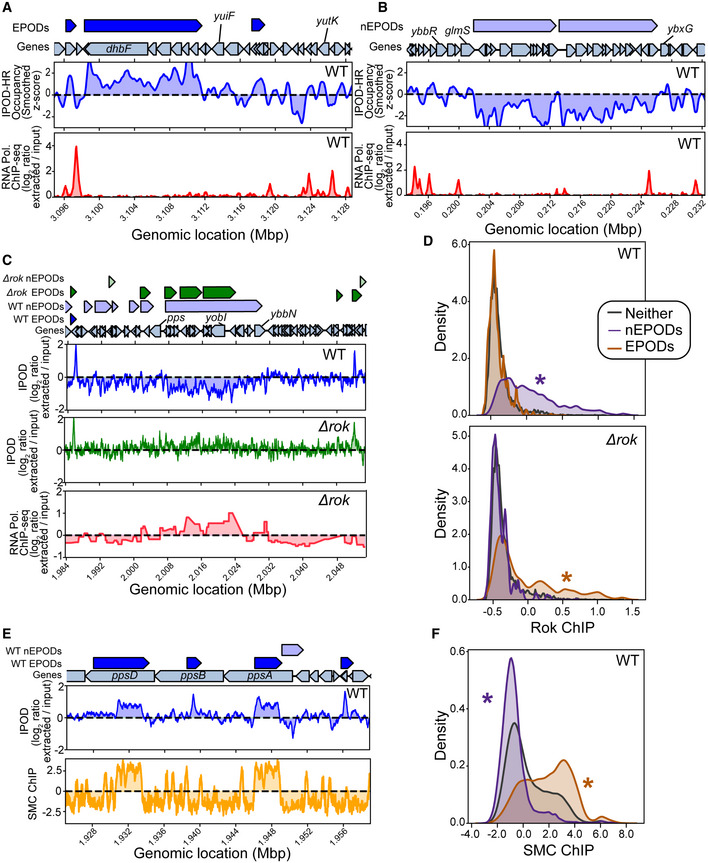
Figure 7. IPOD‐HR in Bacillus subtilis reveals Rok‐bound and SMC‐bound domains.
-
AIPOD‐HR ChIP‐subtracted z scores and RNA polymerase ChIP in the vicinity of an extended protein occupancy domain (EPOD). ChIP tracks are shown as log2 extracted/input ratios; the z score is the ChIP‐subtracted robust z score smoothed with a 512‐bp rolling median.
-
BIPOD‐HR and RNA polymerase ChIP tracks in the vicinity of a negative EPOD (nEPOD).
-
CEffects of deletion of rok in the vicinity of an nEPOD; Rok ChIP data from (Smits & Grossman, 2010) shows a strong overlap with the nEPOD boundary, whereas that occupancy region is lost in Δrok cells.
-
DDistributions of Rok ChIP occupancies (see Materials and Methods) in the EPODs and nEPODs called in WT (top) or Δrok (bottom) cells; note that the Rok ChIP occupancy was taken only in WT cells. (*) indicates a significant difference from the “neither” distribution (that is, genomic sites that are not in an EPOD or nEPOD); P < 0.05, permutation test.
-
EComparison of IPOD occupancy and SMC ChIP occupancy (see Materials and Methods) in the vicinity of several typical EPODs.
-
FGenome‐wide distributions of SMC binding in EPODs versus nEPODs as assessed in WT cells; (*) indicates a significant difference from the background distribution as in panel D.
