ABSTRACT
Bacteria have to process several levels of gene regulation and coordination of interconnected regulatory networks to ensure the most adequate cellular response to specific growth conditions. Especially, expression of complex and costly fitness and pathogenicity-associated traits is coordinated and tightly regulated at multiple levels. We studied the interconnected regulation of the expression of the colibactin and yersiniabactin polyketide biosynthesis machineries, which are encoded by two pathogenicity islands found in many phylogroup B2 Escherichia coli isolates. Comparative phenotypic and genotypic analyses identified the BarA-UvrY two-component system as an important regulatory element involved in colibactin and yersiniabactin expression. The carbon storage regulator (Csr) system controls the expression of a wide range of central metabolic and virulence-associated traits. The availability of CsrA, the key translational regulator of the Csr system, depends on BarA-UvrY activity. We employed reporter gene fusions to demonstrate UvrY- and CsrA-dependent expression of the colibactin and yersiniabactin determinants and confirmed a direct interaction of CsrA with the 5′ untranslated leader transcripts of representative genes of the colibactin and yersiniabactin operons by RNA electrophoretic mobility shift assays. This posttranscriptional regulation adds an additional level of complexity to control mechanisms of polyketide expression, which is also orchestrated at the level of ferric uptake regulator (Fur)-dependent regulation of transcription and phosphopantetheinyl transferase-dependent activation of polyketide biosynthesis. Our results emphasize the interconnection of iron- and primary metabolism-responsive regulation of colibactin and yersiniabactin expression by the fine-tuned action of different regulatory mechanisms in response to variable environmental signals as a prerequisite for bacterial adaptability, fitness, and pathogenicity in different habitats.
KEYWORDS: secondary metabolite, cytopathic effect, BarA-UvrY, two-component regulatory systems, pathogenicity islands, high pathogenicity island
INTRODUCTION
A variety of Escherichia coli strains belonging to the phylogenetic group B2, which comprises extraintestinal pathogenic E. coli (ExPEC) as well as commensal E. coli strains, have been identified to carry two potentially linked genomic islands, namely, the so-called “high pathogenicity island” (HPI) and the polyketide synthase (pks) island. Both islands are chromosomally located in close proximity, and the pks island always coexists with the HPI (1–3). Each island carries all necessary genes for a combined polyketide synthase (PKS) and nonribosomal peptide synthase (NRPS) biosynthesis machinery, producing either colibactin or yersiniabactin.
Yersiniabactin, a metallophore involved in metal homeostasis, was first discovered in Yersinia enterocolitica but was later shown to exist in members of Enterobacterales, such as E. coli (1, 4–7). Yersiniabactin can complex Fe(III) ions and Cu(II) ions and afterward internalize these complexes in a FyuA-dependent manner (8–11). FyuA is an HPI-encoded outer membrane protein, playing an important role in the virulence of ExPEC (12–19). Investigation of the role of the HPI in E. coli gene regulation also indicated a connection between motility and yersiniabactin expression, suggesting the interplay between the flagellar regulatory system and the promoter region of the major HPI regulator YbtA (15, 20, 21).
The pks island encodes the synthesis machinery required for colibactin production. Colibactin is a hybrid polyketide/nonribosomal peptide causing DNA damage in mammalian cells upon infection (22). Different colibactin structures have been suggested recently (23–26). Colibactin mediates cross-linking of DNA, in vitro as well as in vivo, resulting in DNA double-strand breaks, thus provoking cell cycle arrest and inducing megalocytosis/senescence (22, 25–27). This genotoxin promotes bacterial virulence (28, 29), but the ability to produce colibactin has also been correlated with probiotic and analgesic effects (30, 31), potential antimicrobial activity (32), and cancer propagation (33–37). We are also beginning to better understand the extent to which growth conditions and bacterial factors affect the regulation of colibactin expression in E. coli (38–41). To further investigate the regulation of expression, we screened pks island-positive E. coli for their ability to produce colibactin.
Generally, each PKS and/or NRPS machinery-encoding determinant includes its own phosphopantetheinyl transferase (PPTase) gene. However, in E. coli, the HPI lacks a gene coding for a PPTase, and it has been demonstrated that the pks island-encoded PPTase ClbA can also activate PKSs and NRPSs involved in yersiniabactin synthesis (42, 43). Moreover, gene expression of both the HPI and the pks island is regulated by the global iron-responsive regulatory protein ferric uptake regulator (Fur) (44, 45). It was also demonstrated that the E. coli heat shock protein HtpG is required for colibactin and yersiniabactin production (46). The co-occurrence and physical linkage of the pks island and the HPI, together with their common Fur-dependent regulation, the role of the HtpG chaperone for the production of colibactin and yersiniabactin, and the importance of the pks island-encoded PPTase ClbA for yersiniabactin expression demonstrate the close relationship between these two islands coding for secondary metabolites contributing to the fitness of extraintestinal pathogenic and commensal E. coli. Against this background, the findings that yersiniabactin expression is affected by motility regulation (15) and that uropathogenic E. coli (UPEC) model strain 536 and its spontaneous nonhemolytic pathogenicity island I and II deletion mutant 536-21 (47) appeared to be phenotypically colibactin negative despite carrying an intact pks island led us to further investigate the regulation of colibactin and yersiniabactin expression. We applied reporter gene constructs and electrophoretic mobility shift assays, as well as different phenotypic analyses in different strain backgrounds, to demonstrate the complex dependencies inside this regulatory network.
RESULTS
The response regulator UvrY is indispensable for the induction of a colibactin-mediated cytopathic effect.
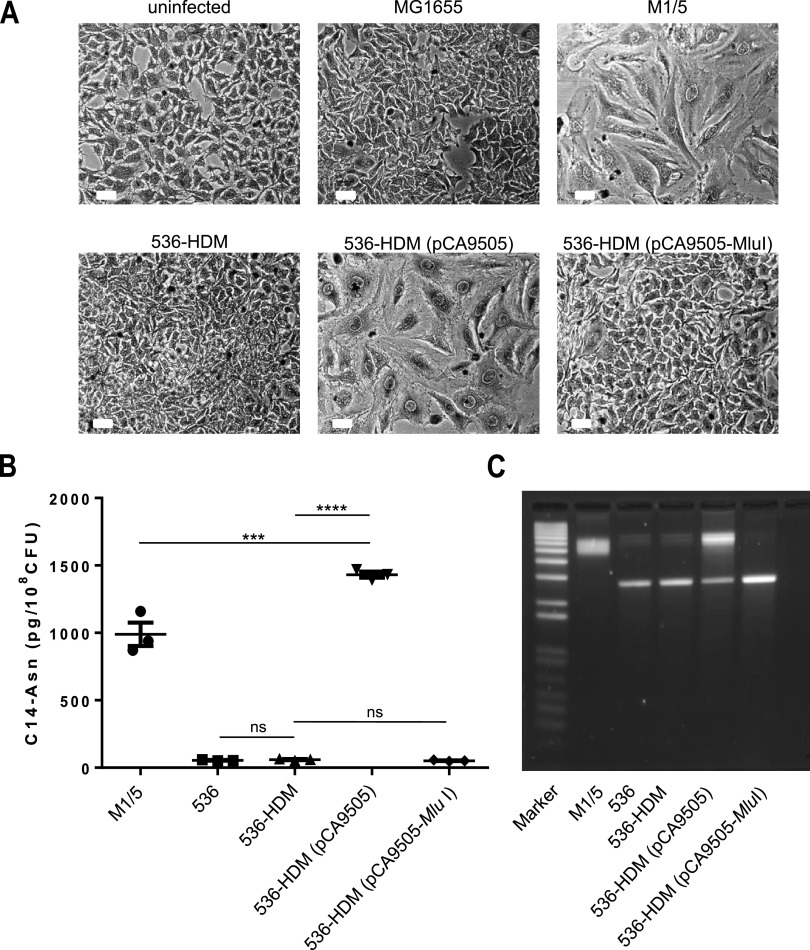
Megalocytosis assays with the nonhemolytic mutant of phylogroup B2 UPEC strain E. coli 536, designated 536-HDM (48), demonstrated that despite the presence of an intact pks island, a cytopathic effect (CPE) could not be observed upon infection (Fig. 1A). As colibactin is first synthetized as a prodrug (so-called precolibactin), carrying an N-myristoyl-d-asparagine (C14-asparagine [C14-asn]) side chain which is cleaved in the periplasm to release the active genotoxin, C14-asn can be used as a readout for colibactin expression. Interestingly, E. coli strains 536 and 536-HDM displayed a low level of C14-asn production as well as a weak DNA cross-link activity (Fig. 1B and C). This finding suggests that the pks island is functional but only weakly expressed in E. coli strain 536. Accordingly, E. coli 536-HDM infected HeLa cells showed a similar cell size as noninfected HeLa cells or HeLa cells incubated with E. coli K-12 strain MG1655 (Fig. 1A), whereas HeLa cell infection with pks-positive phylogroup B2 E. coli isolate M1/5 led to megalocytosis (Fig. 1A) and high levels of N-myristoyl-d-asparagine production, as well as a significantly stronger DNA cross-link activity (Fig. 1B and C). A genome sequence analysis of E. coli strain 536 revealed that the pks island was fully conserved in this strain, but we detected, in comparison to the published colibactin-producing newborn meningitis isolate IHE3034 as well as to K-12 laboratory strain MG1655, a 6.1-kb chromosomal deletion between uvrC and fliY (see Fig. S1 in the supplemental material), which also includes the uvrY gene encoding the response regulator UvrY of the BarA-UvrY two-component regulatory system (TCS) (49). We, therefore, assumed that the presence of UvrY is crucial for the colibactin-mediated phenotypic effect. To test this hypothesis, uvrY plasmid pCA9505 (50) was used to complement the uvrY deletion in E. coli strain 536-HDM. Infection with this complemented strain led to the induction of megalocytosis in HeLa cells, high C14-asn production, and DNA cross-linking activity (Fig. 1A to C) confirming our initial assumption that strain 536 codes for a functional colibactin biosynthesis machinery, which requires UvrY for its proper expression. Transformation of E. coli 536-HDM with a vector control (pCA9505-MluI) did not cause megalocytosis upon HeLa infection. Also, the production of C14-asn and DNA cross-linking activity were not increased compared with those of wild-type strain 536 and its nonhemolytic mutant 536-HDM (Fig. 1A to C).
FIG 1.
The pks island-positive nonhemolytic mutant E. coli 536-HDM requires exogenous UvrY to induce a cytopathic effect in HeLa cells. (A) HeLa cells were either not infected or infected with the indicated E. coli strains to a multiplicity of infection (MOI) of 200. After 4 hours of infection, HeLa cells were washed to remove bacteria and further cultivated. At 72 h postinfection, cells were washed and Giemsa stained. Scale bars, 50 μm. The uvrY plasmid pCA9505 (50) was used to complement the uvrY deletion in E. coli strain 536-HDM. As a control, E. coli strain 536-HDM was transformed with the uvrY-negative variant of plasmid pCA9505, namely, pCA9505-MluI (90). (B) N-Myristoyl-d-asparagine (C14-asparagine) quantification (mean values ± SEM) in bacterial cultures of E. coli strains M1/5, 536, 536-HDM, 536-HDM (pCA9505), and 536-HDM (pCA9505-MluI) by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The data presented in the graph were obtained from three biological replicates. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; 1-way ANOVA with Bonferroni correction. (C) DNA cross-link formation of plasmid DNA exposed to strains M1/5, 536, 536-HDM, 536-HDM (pCA9505), and 536-HDM (pCA9505-MluI) was visualized after migration under alkaline denaturing conditions. M, DNA size marker (1 kb plus DNA ladder, Invitrogen). This image is representative of three independent experiments.
Comparison of the genomic region comprising uvrY in different E. coli strains. Analysis of the chromosomal region next to uvrY in the complete genome sequences of pks island-positive, but CPE-negative, uropathogenic E. coli strain 536 with those of colibactin-producing newborn meningitis isolate IHE3034 as well as to K-12 laboratory strain MG1655 that indicated that E. coli 536 carries a 6.1-kb chromosomal deletion spanning the region between uvrC and fliY and thus lacks the uvrY gene. Download FIG S1, PDF file, 0.2 MB (215.3KB, pdf) .
Copyright © 2022 Rehm et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
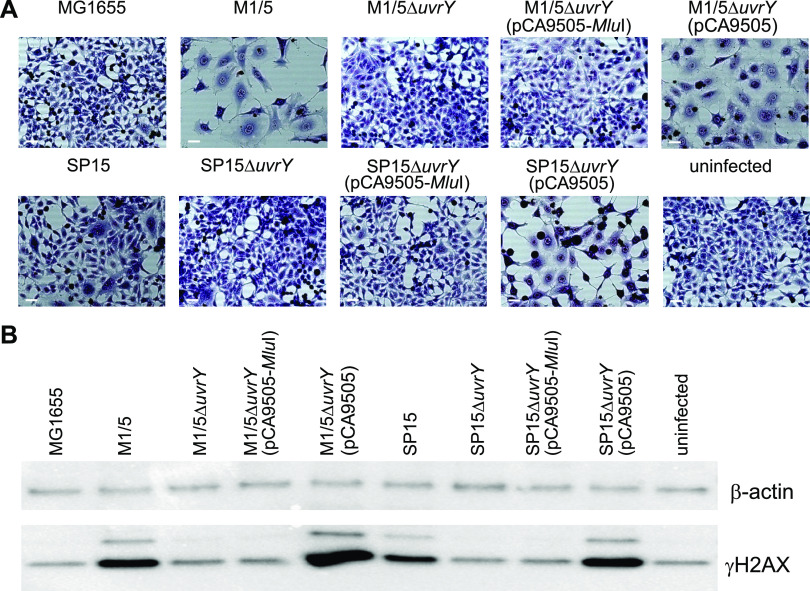
Next, we wanted to investigate if the UvrY dependency of the colibactin-mediated phenotype was restricted to E. coli strain 536. As shown in Fig. 2A, HeLa cell infection with pks-positive E. coli wild-type strains M1/5 and SP15 resulted in strong megalocytosis. Moreover, infection with these strains led to the formation of high γH2AX levels as an indicator of DNA double-strand breaks in mammalian cells (Fig. 2B). As described above, colibactin induces DNA double-strand breaks, which are accompanied by the phosphorylation of the small histone H2AX at serine residue 139. However, infection with uvrY deletion mutants E. coli M1/5ΔuvrY and SP15ΔuvrY abrogated the CPE since no megalocytosis and only low levels of γH2AX could be detected that were comparable to HeLa cell mock infection or infection with E. coli K-12 strain MG1655. Transformation of E. coli strains M1/5ΔuvrY and SP15ΔuvrY with plasmid-encoded uvrY using pCA9505 led to the reconstitution of the CPE as indicated by the appearance of strong γH2AX signals and detection of enlarged cells. In contrast, transformation with the uvrY-negative variant pCA9505-MluI did not lead to the reconstitution of the CPE. Taken together, these results clearly show that, although the colibactin determinant is generally expressed at a very low level, the presence of UvrY is essential for efficient colibactin expression.
FIG 2.
UvrY is indispensable for the efficient induction of double-strand breaks and megalocytosis by pks-positive E. coli strains. (A) HeLa cells were either infected with the indicated E. coli strains (MOI of 200) or not infected. After 4 hours of infection, HeLa cells were washed to remove bacteria and further cultivated. At 72 h postinfection, cells were washed and Giemsa stained. Scale bars, 50 μm. (B) At 8 h postinfection, cells were washed with phosphate-buffered saline (PBS) and lysed. An amount of 5 μg total protein per lane of indicated samples were analyzed by SDS-PAGE and afterward transferred onto a polyvinylidene difluoride (PVDF) membrane. γH2AX was detected using the anti-gammaH2A.X (phospho S139) antibody (Abcam). β-Actin served as a loading control.
The carbon storage regulator (Csr) system is required for the colibactin-mediated cytopathic effect.
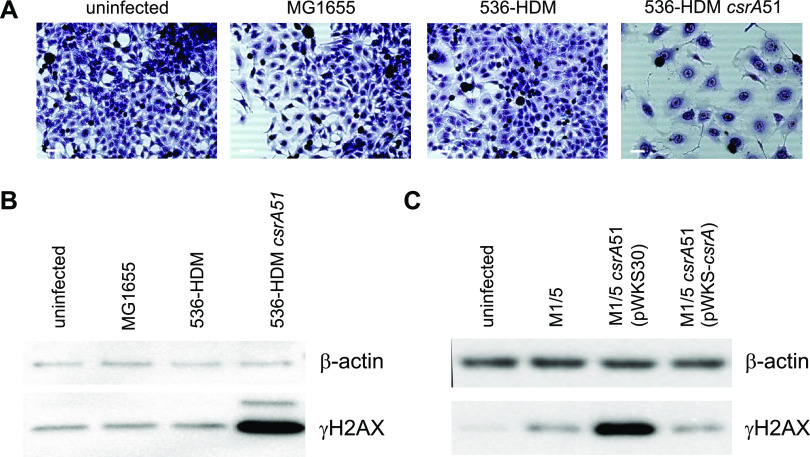
The BarA-UvrY TCS regulates the synthesis of the small noncoding RNAs csrB and csrC, which antagonize the function of the global RNA-binding protein CsrA (51). Since UvrY activates csrB and csrC transcription, we hypothesized that deletion of csrB and csrC would also lead to abrogation of the CPE. Several deletion mutants as well as the corresponding complemented mutants of newborn meningitis E. coli isolate SP15 were generated and tested for their ability to cause megalocytosis in HeLa cells. This E. coli strain has been described previously as evoking colibactin-dependent CPE in mammalian cells (22). Single deletion of either csrB or csrC in E. coli strain SP15 did not lead to an abrogation of the cytotoxic effect, whereas the double deletion mutant SP15ΔcsrBΔcsrC was not able to cause megalocytosis in HeLa cells, indicating that at least one of the two noncoding RNAs is required for proper expression of the colibactin-mediated megalocytosis phenotype. Transformation of this strain with plasmid pRS-csrB reconstituted its ability to cause a CPE (see Fig. S2 in the supplemental material; Table 1). The observation that the csrB and csrC small noncoding regulatory RNAs can complement each other’s function has already been described (52, 53). Accordingly, the regulatory RNAs csrB and csrC regulated by the BarA-UvrY TCS are required in pks-positive strains to evoke a CPE on mammalian cells. The csrB and csrC RNAs sequester and thus antagonize the function of the key translational regulator CsrA (54). To test whether colibactin synthesis and/or delivery was repressed by CsrA, we partially deleted csrA in E. coli strain 536-HDM resulting in the production of a truncated CsrA protein, CsrA51, with reduced functionality (54). While HeLa cells infected with E. coli strains MG1655 or 536-HDM behaved like noninfected cells and neither led to megalocytosis (Fig. 3A) nor γH2AX formation (Fig. 3B), incubation of HeLa cells with the partial csrA deletion mutant E. coli 536-21csrA51 led to the detection of γH2AX as well as megalocytotic cells (Fig. 3A). The same effect was detected upon csrA truncation and complementation of this mutant in E. coli strain M1/5 (Fig. 3C). Thus, CsrA is a repressor of colibactin synthesis and/or the colibactin-mediated CPE.
TABLE 1.
Effect of csrB and crsC deletions in E. coli strains on the colibactin-mediated CPE in HeLa cells
| Strain | Megalocytosis phenotype |
|---|---|
| SP15 | + |
| SP15 ΔcsrB | + |
| SP15 ΔcsrC | + |
| SP15 ΔcsrB ΔcsrC | − |
| SP15 ΔcsrB ΔcsrC pRS-csrB | + |
FIG 3.
CsrA blocks the colibactin-mediated cytopathic effect of pks-positive E. coli strains. (A) HeLa cells were either not infected or infected with the indicated E. coli strains (MOI of 200). After 4 hours of infection, HeLa cells were washed to remove bacteria and further cultivated. At 72 h postinfection, cells were washed and Giemsa stained. Scale bars, 50 μm. Eight hours postinfection, cells were washed with PBS and lysed. A total protein amount of 5 μg (B) or 4 μg (C) per lane of indicated samples was analyzed by SDS-PAGE and afterward transferred onto PVDF membranes. γH2AX was detected using either an anti-gammaH2A.X (phospho S139) antibody (Abcam) (B) or anti-phospho-Histone H2A.X (Ser139), clone EP854(2)Y (Merck-Millipore) (C). β-Actin served as a loading control.
Impact of the small regulatory RNAs csrB and csrC on the cytopathic effect of E. coli strain SP15 in HeLa cells. HeLa cells were either not infected or infected with the indicated E. coli strains to a multiplicity of infection (MOI) of 250. After 4 hours of infection, HeLa cells were washed to remove bacteria and further cultivated. At 72 h postinfection, cells were washed and Giemsa stained. Scale bars, 100 μm. Plasmid pRS-csrB was used to complement the csrB and csrC deletion in E. coli strain SP15ΔcsrB ΔcsrC. Download FIG S2, PDF file, 0.4 MB (382.3KB, pdf) .
Copyright © 2022 Rehm et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CsrA directly represses the synthesis of enzyme(s) encoded by the pks island.
Since we demonstrated in different E. coli strain backgrounds that CsrA is involved in the colibactin synthesis and/or the colibactin-mediated host cell damage, we investigated whether CsrA directly represses the expression of colibactin genes. A SELEX-derived consensus motif from 55 ligands for E. coli CsrA binding has been published (55, 56). The motif was determined as RUACARGGAUGU, with the underlined sequence being 100% conserved and the R representing a purine base. When we performed a bioinformatics search for respective motifs covering the nucleotide sequence ACARGGA within the 19 genes of the pks island of E. coli M1/5, we detected 7 motifs of putative CsrA binding sequences (Table 2).
TABLE 2.
Sequences in the pks island with a putative CsrA binding motif
| Location | Sequencea |
|---|---|
| Within clbG | GAACAGGGATTT |
| Within clbI | TCACAGGGACGT |
| Within clbJ | TCACAGGGATGT |
| Directly upstream of clbL | CAACAGGGAGAA |
| Within clbN | TGACAAGGAGAA |
| Directly upstream of clbQ | CAACAAGGAGTG |
| Directly upstream of clbS | ATACAAGGAGCA |
Nucleotides, which are 100% conserved in the consensus motif described by Dubey and colleagues, (55) are underlined.
The clbQ gene was among the genes comprising a putative CsrA binding motif in its 5′ untranslated leader sequence (Fig. 4A). ClbQ is a thioesterase and one of the last enzymes of the colibactin production machinery responsible for the hydrolytic cleavage of precolibactin from polyketides. We examined a putative interaction of in vitro-transcribed clbQ RNA with CsrA in an RNA electrophoretic shift assay (RNA EMSA). When 3′ biotin-labeled clbQ RNA was incubated with increasing amounts of purified CsrA protein, a clear shift of the RNA band was observed (Fig. 4B). The addition of an excess amount of unlabeled clbQ RNA as a competitor reverted the band shift but not 100-fold excess of phoB RNA, which was used as a negative control (57) (Fig. 4C). These results suggest a specific and direct interaction of CsrA with the clbQ untranslated leader RNA in vitro.
FIG 4.
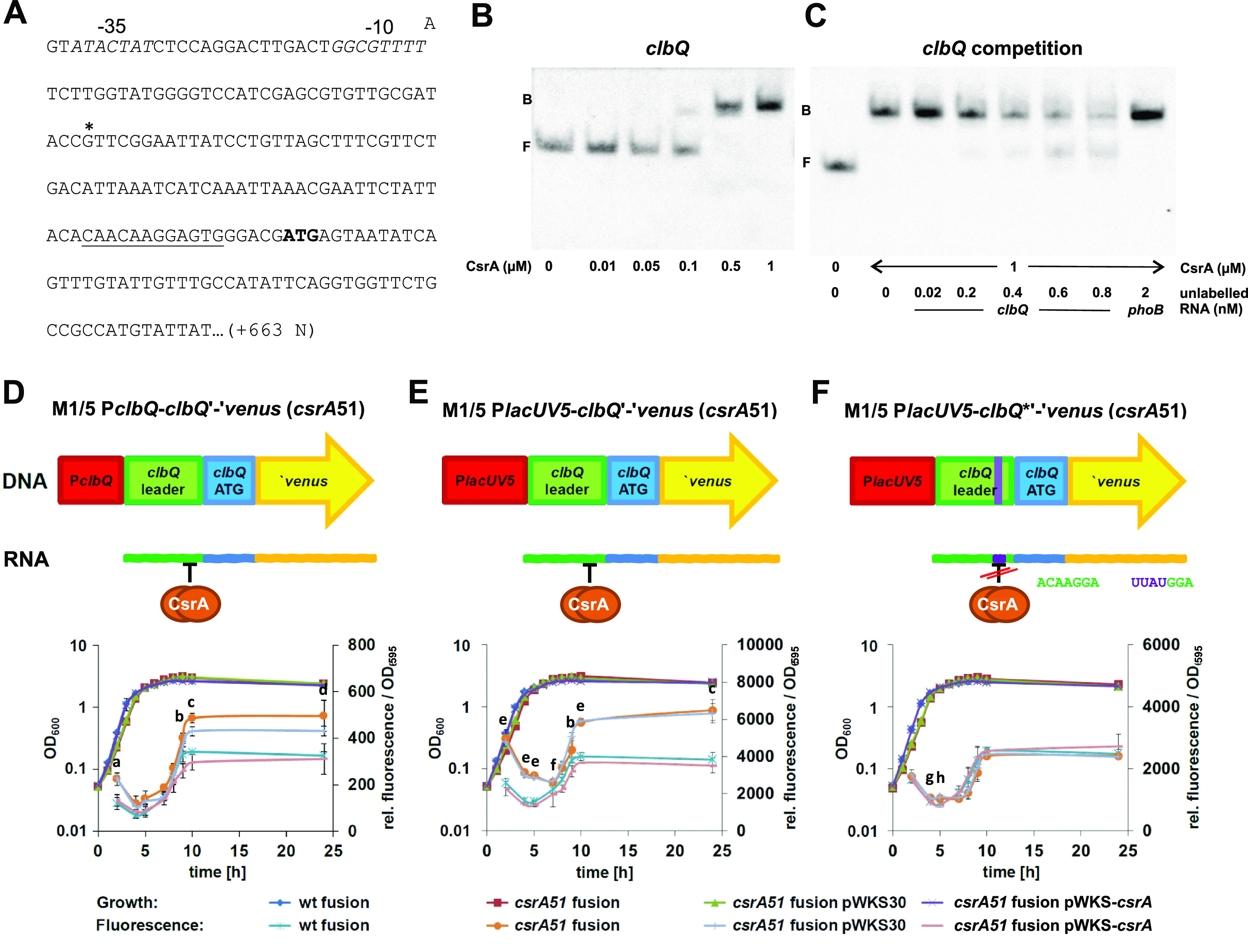
Expression of clbQ is directly repressed by CsrA in E. coli strain M1/5. (A) The partial sequence of the clbQ locus is shown, including the −35 and −10 regions (in italics) and the transcriptional start (*). The putative CsrA-binding motif is underlined, and the start codon for translation is indicated in bold. The sequence from the transcription start downstream corresponds to the sequence used for clbQ RNA probe generation used in RNA electrophoretic mobility shift assays (EMSAs) shown in Fig. 4B and C. (B) An RNA EMSA with a biotin-labeled clbQ RNA probe (20 pM) and increasing amounts of purified CsrA protein (10 nM, 50 nM, 100 nM, 500 nM, and 1 μM; F, free probe; B, bound probe) was performed. The sequence from the transcription start corresponds to the sequence used for clbQ RNA probe generation. (C) Biotin-labeled clbQ RNA probe (20 pM) was incubated with 1 μM CsrA and increasing amounts of unlabeled clbQ RNA (20 pM, 200 pM, 400 pM, 600 pM, and 800 pM; f.p., free probe) or phoB RNA (2 nM) in a competitive RNA EMSA. (D to F) Growth and fluorescence of a set of E. coli M1/5 versus E. coli M1/5 csrA51 reporter gene fusion strains with venus as a reporter gene in a translational fusion (D), with the lacUV5 promoter instead of the native clbQ promoter (E), and with the lacUV5 promoter containing a modified sequence within the putative CsrA-binding motif (F) were monitored in M9 minimal medium without glucose but containing pyruvate and casein hydrolysate for 24 h at the indicated time points. Means and standard deviations of three independent experiments are shown for the fluorescence values. Growth curves of only one experiment are depicted since growth was essentially the same in all three experiments. Statistical analyses using the 1-way ANOVA test were performed comparing the fluorescence of E. coli M1/5 PclbQ-clbQ'-'venus to E. coli M1/5 csrA51 PclbQ-clbQ'-'venus (group 1) and E. coli M1/5 csrA51 PclbQ-clbQ'-'venus pWKS30 to E. coli M1/5 csrA51 PclbQ-clbQ'-'venus pWKScsrA (group 2) for each time point. Small letters in D, E, and F correspond to significance values of group1/group 2 as follows: a, P < 0.01/P < 0.01; b, ns/P < 0.01 (b); c, P < 0.001/P < 0.001; d, P < 0.01/P < 0.05; e, P < 0.0001/P < 0.0001; f, P < 0.01/P < 0.0001; g, P < 0.05/P < 0.01; h, P < 0.05/P < 0.05. If not marked, no significance was found.
To test whether CsrA repressed clbQ expression, a set of reporter gene fusions were generated in E. coli strain M1/5 using the venus gene as a reporter. The clbQ locus was chromosomally manipulated either in the E. coli M1/5 wild type or in the mutant strain E. coli M1/5 csrA51 with a less functional CsrA protein. The growth and fluorescence of the resulting reporter strains were examined for 24 h. Fluorescence of the translational clbQ-venus fusion strain with a truncated CsrA protein, E. coli M1/5 PclbQ-clbQ'-'venus csrA51, was elevated about 1.6-fold compared with that of the wild-type strain M1/5 PclbQ-clbQ'-'venus after 10 h of growth (Fig. 4D). Bacterial growth remained almost unaffected by csrA manipulation throughout the experiment. However, complementation with plasmid-encoded CsrA using pWKS-csrA, but not with empty vector control pWKS30, resulted in fluorescence comparable to the wild type. These results confirmed that CsrA represses venus (clbQ) expression. A similar effect was observed when the clbQ promoter of the translational clbQ-venus fusion strains was exchanged by the constitutive lacUV5 promoter (Fig. 4E), where venus fluorescence of E. coli M1/5 PlacUV5-clbQ'-'venus csrA51 was also increased 1.6-fold compared with that of M1/5 PlacUV5-clbQ'-'venus after 10 h of growth. Therefore, repression of venus expression by CsrA seemed to be independent of the clbQ promoter but was probably attributed to the presence of the predicted CsrA binding site in the clbQ leader sequence. To further test this hypothesis, the sequence encoding the putative CsrA binding motif of the reporter construct with the lacUV5 promoter was genetically modified from CAACAAGGAGTG to CATTATGGAGTG. The resulting strains M1/5 PlacUV5-clbQ*'-'venus and M1/5 PlacUV5-clbQ*'-'venus csrA51 exhibited similar fluorescence throughout growth (Fig. 4F). Accordingly, alteration of the CsrA binding motif resulted in the abrogation of the inhibitory effect of CsrA on venus expression. We also compared the fluorescence of another set of reporter strains, namely, E. coli strains M1/5 PclbQ-AL-venus and M1/5 PclbQ-AL-venus csrA51, in which the native clbQ promoter was fused to a sequence encoding an artificial 5′ untranslated leader (AL) without the CsrA binding motif (see Fig. S3A in the supplemental material) after 10 h of growth. Fluorescence of both strains was comparable (Fig. S3B), confirming again that CsrA did not influence clbQ promoter activity and acted only on the 5′ untranslated clbQ leader sequence. Taken together, we demonstrate that CsrA represses clbQ expression directly by binding to the clbQ untranslated RNA leader.
Activity of the clbQ promoter fused to a sequence encoding an artificial 5′ untranslated leader and venus as a reporter gene. (A) Sequences encoding the clbQ 5′ leader and the artificial leader are shown in italics, an asterisk represents the transcription start, and the translational start codon is depicted in bold. The sequence encoding the putative CsrA-binding motif is underlined. (B) Expression from the clbQ promoter in E. coli strains M1/5 PclbQ-AL-'venus and M1/5 PclbQ-AL-'venus was measured by means of venus fluorescence after 10 h of growth in M9 minimal medium without glucose but containing pyruvate and casein hydrolysate. Mean values with standard deviations of three independent experiments are shown. Download FIG S3, PDF file, 0.2 MB (248.3KB, pdf) .
Copyright © 2022 Rehm et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The Csr system influences yersiniabactin levels.
Coregulation of the expression of two probably coselected fitness determinants makes sense, and consequently, we assumed that the Csr system is involved not only in the regulation of the pks island-encoded polyketide colibactin but also in that of the HPI-encoded polyketide yersiniabactin.
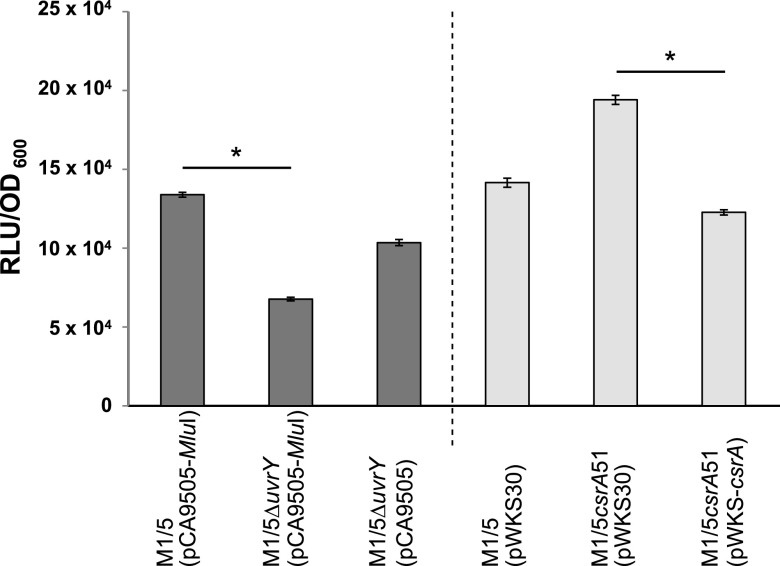
In the absence of UvrY, pks-positive E. coli isolates were not able to cause the cytopathic effect on HeLa cells, and we showed that CsrA was responsible for the repression of colibactin expression. To investigate the influence of UvrY and CsrA on yersiniabactin expression, we employed a luciferase reporter system to indirectly quantify yersiniabactin levels produced by E. coli strain M1/5 carrying or lacking either uvrY or a full-length csrA gene, respectively. Since yersiniabactin is synthesized only when iron availability is limited, quantification was performed with bacteria grown under iron-limiting conditions by adding 2,2′-dipyridyl as an iron-chelating agent. As shown in Fig. 5, yersiniabactin levels in E. coli strain M1/5 ΔuvrY carrying pCA9505-MluI were significantly reduced compared with those in its uvrY-positive counterpart E. coli M1/5 (pCA9505-MluI). Transformation of the uvrY deletion mutant with the uvrY-harboring plasmid pCA-9505 resulted in the production of yersiniabactin in amounts almost comparable to that of the wild-type strain with the empty plasmid, namely, E. coli M1/5 (pCA9505-MluI). Because of the negative influence of CsrA on colibactin production, we expected a similar effect of CsrA on yersiniabactin biosynthesis. Indeed, E. coli strain M1/5 csrA51 (pWKS30), which expresses the truncated and thus less functional CsrA protein, produced more yersiniabactin than E. coli strain M1/5 (pWKS30). When strain M1/5 csrA51 was complemented with the csrA-harboring plasmid pWKS-csrA, yersiniabactin synthesis was restored to the wild-type level. Thus, we show that CsrA represses phenotypic expression not only of colibactin but also of yersiniabactin.
FIG 5.
CsrA and UvrY affect yersiniabactin levels in E. coli strain M1/5. Yersiniabactin levels were quantified in the supernatants of indicated E. coli M1/5 strains grown for 24 h under iron-limiting conditions using Salmonella reporter strain WR1542. The means of three experiments are shown with standard deviations. RLU, relative light units; *, P < 0.05.
CsrA directly represses the synthesis of YbtA encoded by the high pathogenicity island.
We assumed that CsrA might directly repress the production of proteins required for yersiniabactin synthesis. Therefore, we screened for putative CsrA binding motifs within the HPI as described for the colibactin-encoding pks island using the 100% conserved binding motif sequence ACARGGA (55). As indicated in Table 3, we detected seven sites comprising putative CsrA binding sequences within the HPI. In the case of ybtA, this motif is located in their 5′ untranslated leader sequences. The ybtA upstream region, including a putative CsrA binding motif and two YbtA binding sites, is shown in Fig. 6A. First, we investigated by RNA EMSA whether the 5′ ybtA leader sequence was bound by purified CsrA. The ybtA probe comprising the sequence from the ybtA transcription start as well as the YbtA and Fur binding sites (Fig. 6A) showed a clear retardation of migration in RNA EMSA upon incubation with increasing amounts of purified CsrA protein (Fig. 6B). The addition of excess unlabeled ybtA RNA to compete for CsrA binding led to a marked reduction of the shift, whereas the addition of 100-fold excess phoB RNA (negative control) did not reduce the RNA shift (Fig. 6C).
TABLE 3.
Sequences in the high pathogenicity island with a putative CsrA-binding motif
| Location | Sequencea |
|---|---|
| Within ybtS | AAACAAGGATGC |
| Directly upstream of ybtA | CCACAGGGAGAT |
| Within irp2 | CCACAAGGACAA |
| Within irp1 | CGACAAGGATGG |
| Within ybtT | CCACAAGGACTG |
| Directly upstream of fyuA | TTACAGGGACTC |
| Within fyuA | CCACAGGGAACG |
Nucleotides, which are 100% conserved in the consensus motif described by Dubey and colleagues (55), are underlined.
FIG 6.
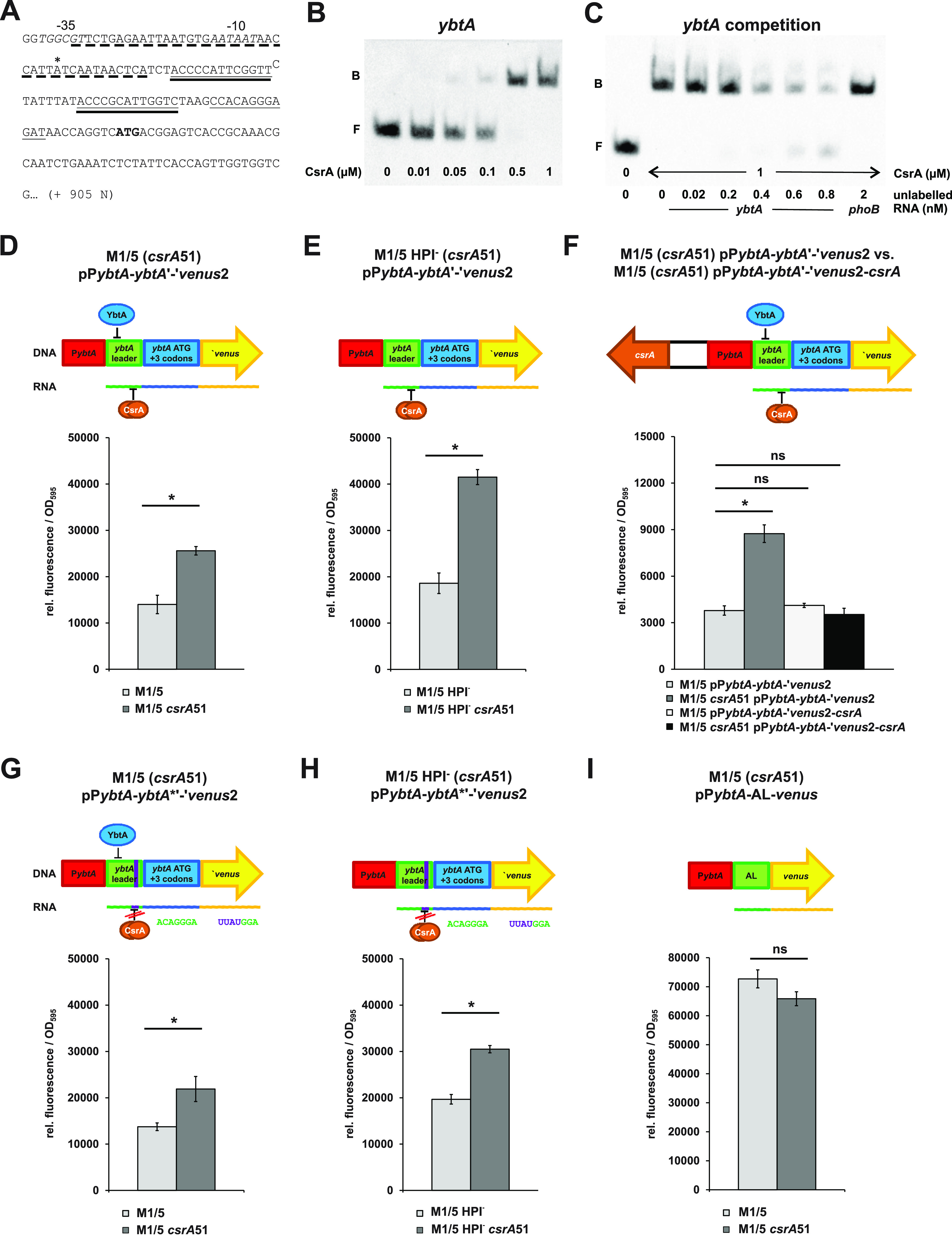
CsrA affects the expression of ybtA in E. coli strain M1/5. (A) Partial sequence of the ybtA locus with its −35 and −10 regions (in italics) and the transcriptional start site (*). Nucleotides comprising the Fur-binding motif are underlined by a dotted line, whereas YbtA-binding sites are underlined twice. The sequence encoding the putative CsrA-binding motif is highlighted by a continuous line and the translation start codon is shown in bold. The sequence depicted starting from the transcription start was used for ybtA RNA probe generation used in RNA EMSA experiments. (B) The biotin-labeled ybtA RNA probe (20 pM) was incubated without or with increasing amounts of purified CsrA protein (10 nM, 50 nM, 100 nM, 500 nM, and 1 μM; F, free probe; B, bound probe). (C) A competitive RNA EMSA was performed with 20 pM of biotin-labeled ybtA RNA probe, 1 μM CsrA, and increasing amounts of unlabeled ybtA RNA (20 pM, 200 pM, 400 pM, 600 pM, and 800 pM; f.p., free probe) or phoB RNA (2 nM). (D to I). Plasmid-based ybtA-venus reporter gene fusions were generated, and the fluorescence (relative fluorescence referred to optical density at 595 nm [OD595]) between csrA-positive E. coli M1/5 and E. coli M1/5 csrA51 background strains after 20 h of cultivation was compared. For this comparison, cells were grown in M9 minimal medium without glucose but containing pyruvate and casein hydrolysate. 2,2′-Dipyridyl was added to relieve Fur-mediated repression of the yersiniabactin operon. D and E show fluorescence of E. coli strains M1/5 and M1/5 csrA51 or E. coli M1/5 HPI− and M1/5 HPI− csrA51, respectively, when transformed with a plasmid carrying a translational ybtA-venus fusion with the native ybtA promoter and leader sequence. (F) Fluorescence of E. coli strains M1/5 and M1/5 csrA51 transformed with plasmids containing the translational ybtA-venus fusion only, pPybtA-ybtA'-'venus2, or carrying a csrA locus in addition for complementation, pPybtA-ybtA'-'venus2-csrA. G and H show the fluorescence of E. coli strains M1/5 and M1/5 csrA51 or E. coli M1/5 HPI− and M1/5 HPI− csrA51, respectively, when transformed with a plasmid carrying the translational ybtA-venus fusion with a sequence modification leading to a disrupted CsrA-binding motif. (I) Fluorescence of E. coli strains M1/5 and M1/5 csrA51 carrying a transcriptional venus fusion comprising the ybtA promoter fused to a sequence encoding an artificial 5′ leader (AL) and venus. *, P < 0.05; ns, not significant.
Several plasmid-based ybtA-venus reporter fusions were generated to compare ybtA promoter activity in the E. coli M1/5 wild type and its isogenic mutant M1/5 csrA51. When E. coli strains M1/5 and M1/5 csrA51 were transformed with the ybtA translational fusion plasmid pPybtA-ybtA'-'venus2, a significant increase of venus expression could be observed in the mutant expressing the truncated, less functional CsrA protein (Fig. 6D), even though YbtA-mediated repression of the fusion still occurred due to the presence of a functional yersiniabactin-encoding gene cluster. CsrA-dependent repression of ybtA promoter activity was even more pronounced upon relief of YbtA-mediated repression when we compared fluorescence resulting from the same plasmid in E. coli strains M1/5 HPI- and M1/5 HPI- csrA51, which carry a partial deletion of the HPI, including ybtA (Fig. 6E). Introduction of a fully functional csrA gene into pPybtA-ybtA'-'venus2, resulting in plasmid pPybtA-ybtA'-'venus2-csrA, restored functional CsrA expression in E. coli M1/5 csrA51, thus reducing ybtA promoter activity to a similar level as that in the csrA-positive strain M1/5 (pPybtA-ybtA'-'venus2) (Fig. 6F).
Next, we examined if CsrA repressed ybtA promoter activity by direct interaction with the putative CsrA binding motif present in the 5′ untranslated ybtA leader (see Fig. 6A). Similar to our experiments on clbQ expression, the putative CsrA motif CCACAGGGAGAU was therefore genetically modified to CCUUAUGGAGAU, which should interfere with proper CsrA binding. When venus expression was compared in the ybtA-positive (M1/5 versus M1/5csrA51) or ybtA-negative (M1/5 HPI- versus M1/5 HPI-csrA51) strain pairs using plasmid pPybtA-ybtA*'-'venus2, fluorescence was increased in variants expressing the truncated CsrA protein (Fig. 6G or H, respectively). Disorganization of the putative CsrA binding motif in plasmid pPybtA-ybtA*'-'venus2 diminished the fold increase in venus expression due to csrA truncation. Expression of venus increased upon csrA truncation by 1.59-fold (ybtA-positive strain background) and 1.55-fold (ybtA-negative strain background). In contrast, in the presence of an intact YbtA binding site, the reporter gene expression was 1.83-fold (ybtA-positive strains) and 2.23-fold (ybtA-negative strains) higher upon truncation of csrA (data used for calculation based on data shown in Fig. 6D and G and Fig. 6E and H, respectively). Reporter gene assays with the ybtA-venus fusion plasmid pPybtA-AL-venus that carried the ybtA promoter fused to an artificial leader without CsrA binding motif (AL) (see Fig. S2) demonstrated that in the absence of a CsrA binding motif no significant difference in fluorescence was detected between the strains M1/5 and M1/5 csrA51 (Fig. 6I). Accordingly, the ybtA promoter activity was not affected by CsrA itself. Our results show that ybtA expression is repressed by CsrA. This effect is only partially due to the direct interaction of CsrA with the ybtA leader. Consequently, another so far unidentified CsrA-governed mechanism seems to be involved in the control of ybtA expression.
DISCUSSION
The multilayered intertwining of two determinants encoding secondary metabolites with different functions in E. coli is remarkable. Secondary metabolite production is an energy-demanding process that also requires the provision of certain primary metabolic precursors, such as acyl-coenzyme A (CoA) moieties. Tight and coordinated regulation of secondary metabolite production is thus a way to minimize the production costs and to ensure that the bacterial producer can flexibly meet the challenges under different growth conditions and in different habitats. The observation that the pks island is always accompanied by the HPI in phylogroup B2 E. coli strains was made quite soon after the colibactin island was first described (1). It has also been shown that the HPI including flanking chromosomal regions can be horizontally transferred as a larger DNA entity by F-plasmid-mediated mobilization and subsequent recombination (58). Although HPI is not self-transferable, comparative genomic analyses and conjugation experiments indicated that joint horizontal transfer of the HPI can occur along with the pks island and another adjacent chromosomal island (59). In other colibactin-producing coliform enterobacteria, such as Klebsiella pneumoniae, Citrobacter koseri, and Klebsiella (formerly Enterobacter) aerogenes, the colibactin and yersiniabactin determinants are colocalized within an integrative and conjugative element (ICE) (1–3), confirming that physical linkage of the colibactin and yersiniabactin islands is actively supported by joint acquisition or spread via horizontal gene transfer.
The pks island and the HPI are not only physically linked but also interconnected at different regulatory levels of gene expression. The expression of both polyketides responds to iron availability. The ferric uptake regulator (Fur) is involved in the regulation of colibactin and yersiniabactin expression (4, 44, 45, 60). Fur represses the transcription of the small regulatory RNA ryhB (61). As a result, colibactin expression is also regulated at the posttranscriptional level (43, 45). The ryhB regulatory RNA has been shown to repress the expression of the serine acetyltransferase CysE, which facilitates the channeling of serine as a building block into enterobactin synthesis (62). Serine is also a building block of colibactin and yersiniabactin. Accordingly, the effect of ryhB on the expression of colibactin and yersiniabactin is not only due to an iron-dependent regulation of gene expression but also most likely on increased efficiency of nonribosomal peptide biosynthesis. In addition, the intertwining of the biosynthesis pathways of different siderophores, such as enterobactin and yersiniabactin, with colibactin has been demonstrated by showing that the PPTase ClbA, encoded by the colibactin gene cluster, can contribute not only to the synthesis of colibactin but also to the synthesis of yersiniabactin (42). This functional interchangeability of the ClbA PPTase is facilitated by the physical and regulatory linkage of both islands. Another aspect of the interplay between the biosynthesis pathways of the two polyketides is the involvement of the chaperone HtpG and the protease ClpQ. The heat shock protein HtpG appears to protect proteins involved in colibactin and yersiniabactin production from ClpQ-mediated degradation (46).
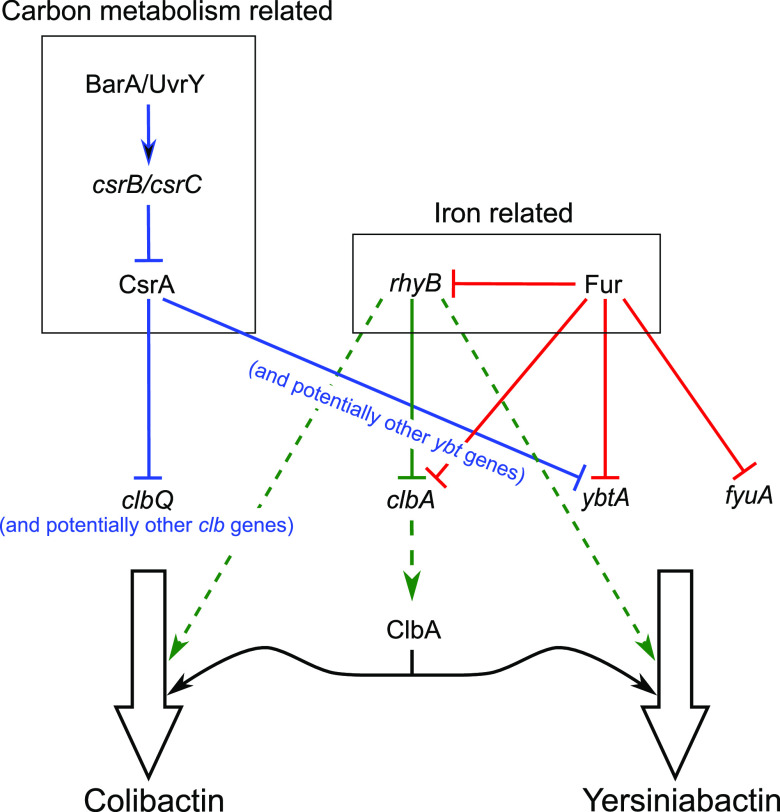
In addition to iron availability, we show that the central metabolic state of the bacterial cells has a strong common impact on colibactin and yersiniabactin expression via direct dependence on the BarA/UvrY two-component system which is integrated into the carbon storage regulator signaling network (Fig. 7) and thus is closely linked to the central carbon metabolism. The sensor kinase BarA senses an appropriate environmental signal and transmits this signal by phosphorylating its cognate response regulator UvrY. Phosphorylated UvrY activates the transcription of the small noncoding RNA (sRNA) loci csrB and csrC. The sRNAs csrB and csrC compete with other RNA targets for CsrA binding, limiting the availability of this RNA-binding repressor (49, 55). CsrA is a global RNA-binding protein and a posttranscriptional regulator. It can either repress or enhance the expression of its RNA targets. CsrA has mostly been described as interacting with a specific sequence within the 5′ untranslated region (5′ UTR) of transcripts, with GGA being the minimal binding sequence (55, 56, 63). CsrA has also been reported to modulate BarA kinase activity as well as BarA-independent regulation of UvrY (49).
FIG 7.
Integration of pks and HPI expression regulation into the Csr- and Fur-dependent regulatory networks. Colibactin and yersiniabactin expression both respond to central carbon- and iron-dependent regulation. CsrA inhibits at the posttranscriptional level expression of several genes of the colibactin and high pathogenicity island. The ferric uptake regulator (Fur) inhibits transcription of clbA, but also of ybtA and fyuA. The small regulatory noncoding RNA ryhB inhibits clbA expression at the posttranscriptional level. The ryhB regulatory RNA also modulates the efficiency of nonribosomal peptide biosynthesis via channeling of serine as a building block into colibactin and yersiniabactin biosynthesis (dashed arrow). Furthermore, the phosphopantetheinyl transferase ClbA activates polyketide synthases and nonribosomal peptide synthetases of both the colibactin as well as the yersiniabactin biosynthesis machinery.
Fine-tuning gene expression is essential for the optimized production of complex metabolites, such as colibactin and yersiniabactin. Here, we demonstrate a direct link between primary and secondary metabolism by showing that the Csr system regulates colibactin and yersiniabactin production at the posttranscriptional level. We show that UvrY is required for the proper phenotypic expression of colibactin and yersiniabactin. Our study also provides evidence that functional CsrA is crucial to keep colibactin and yersiniabactin production in check via posttranscriptional repression of gene expression required for colibactin biosynthesis or regulation of colibactin expression. We have experimentally demonstrated that CsrA represses the expression of the thioesterase ClbQ. The additional presence of the CsrA binding motif in the upstream region of clbL and clbS, as well as in the coding sequence of four additional genes (clbG, clbI, clbJ, and clbN) involved in colibactin biosynthesis (Table 2), suggests that CsrA-dependent posttranscriptional regulation acts at multiple sites of the colibactin determinant. Similarly, we also detected multiple conserved CsrA binding sites in the yersiniabactin gene cluster upstream of ybtA and fyuA as well as within several coding regions (Table 3). Under iron limitation, the AraC-type regulator YbtA is required to initiate the expression of almost all HPI genes, which are subdivided into four transcriptional units (ybtPQXS, ybtA, irp2-irp1-ybtUTE, and fyuA), with the exception of its own gene (20, 64). The expression of ybtA is repressed by the global iron regulator Fur in an iron-rich environment (4, 60, 65, 66). Moreover, ybtA is subject to negative autoregulation (21), and two YbtA binding sites are located in the 5′ untranslated ybtA leader region. FyuA is the yersiniabactin receptor located in the outer membrane and is responsible for the import of iron-charged yersiniabactin. Its synthesis is strongly repressed by Fur and needs to be activated by YbtA (21, 64). Thus, CsrA-dependent posttranscriptional regulation adds another level of complexity to the already diverse checkpoints of colibactin and yersiniabactin expression. In this way, different signals, i.e., iron availability and central metabolic state, are integrated into the multilayered regulation of these fitness and pathogenicity factors.
The Csr system represents an important regulatory mechanism at the posttranscriptional level, which coordinates the expression of specific fitness and pathogenicity-associated traits with relevant physiological conditions. The RNA-binding regulatory protein CsrA is involved in the regulation of various cellular processes, including the synthesis of virulence factors in a wide range of bacteria (51, 67–71). CsrA-dependent regulation of iron uptake and siderophore expression has been reported in several pathogens, including Yersinia pseudotuberculosis, Legionella pneumophila, Salmonella enterica serovar Typhimurium, Pseudomonas aeruginosa, and also in Clostridium acetobutylicum (72–78). CsrA has been described to regulate the expression of the iron uptake system enterobactin in enteropathogenic E. coli (79). It has also been shown that siderophore synthesis and iron transport are regulated in Pseudomonas fluorescens and Vibrio fischeri by the UvrY homologue GacA (80, 81). The observation that the loss of UvrY resulted in a reduced expression of polyketide biosynthesis enzymes in Photorhabdus luminescens accompanied by decreased virulence in insects (82) further confirms that the Csr system plays an important role in controlling the expression of functionally different pathogenicity factors, including iron uptake systems and polyketides. Consequently, the Csr system is generally assumed to link carbon metabolism and iron uptake to optimize fitness during infection. This coupling is critical for the successful colonization or the establishment of an infection because it enables an adequate and fine-tuned modulation of bacterial gene expression in response to individual host environments and associated changes in nutritional demands, e.g., during the course of an infection.
Concerning colibactin and yersiniabactin, we describe a relationship between colocalization and coexpression. This “guilt-by-association” relationship (83) highlights that the encoded polyketides are coexpressed and are required for the same bacterial phenotype or trait. Both polyketides have been described as important fitness and pathogenicity factors in extraintestinal pathogenic E. coli (28, 29, 84, 85). Although different hypotheses regarding the biologically relevant function of colibactin exist, i.e., genotoxin/cyclomodulin versus bacteriocin (86), it is clear that the expression of colibactin and yersiniabactin can promote bacterial growth and survival in vivo, e.g., during infection. The production of these secondary metabolites and their corresponding large biosynthetic machineries also incurs high metabolic costs. As certain fitness factors provide important advantages in certain situations, the metabolic costs associated with them may be disadvantageous in microenvironments, in which they are not required. Accordingly, the precise regulation of gene expression in response to variable environmental signals is a prerequisite for bacterial adaptability, fitness, and pathogenicity in different habitats. This strategy is common in secondary metabolite production, as the complex and interconnected regulation of secondary metabolite expression by pathway-specific as well as global regulatory mechanisms has also been reported for various fungi (87). Against this background, our work emphasizes the importance of the interconnection between iron- and primary metabolism-responsive regulation of colibactin and yersiniabactin expression through the fine-tuned action of transcriptional regulators, such as Fur; posttranscriptional regulators, such as the ryhB sRNA; and the CsrA RNA-binding protein, as well as the posttranslational acyl group activation during colibactin and yersiniabactin biosynthesis via the PPTase ClbA.
MATERIALS AND METHODS
Bacterial strains, plasmids, genetic manipulations, and media.
Bacterial strains and plasmids used in this study are listed in Table 4 and 5, respectively.
TABLE 4.
E. coli strains used in this study
| Strain | Genotype and/or characteristics | Reference |
|---|---|---|
| E. coli | ||
| 536 | pyelonephritis isolate 536; pks+, HPI+ (O6:K15:H31) | 47 |
| 536-HDM | 536ΔhlyI, ΔhlyII::cat | 48 |
| 536-HDM csrA51 | 536-HDM csrA51::cat | This study |
| BL21 (DE3) | F-, gal met r-m-hdsS λlysPlacUV5-T7-Gen1 PlacIqlacI | 91 |
| DH5α | F-endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(argF-lacZYA) U169 (Φ60ΔlacZ M15λ-) | 92 |
| IHE3034 | newborn-meningitis isolate; pks+, HPI+ (O18:K1:H7) | 93 |
| M1/5 | Fecal isolate of a healthy individual; pks+, HPI+, Strr | 42 |
| M1/5 csrA51 | M1/5 csrA153::FRT-kan-FRT; Kanr, Strr | This study |
| M1/5 ΔuvrY | M1/5 ΔuvrY::FRT-cat-FRT; Cmr, Strr | This study |
| M1/5 HPI- | M1/5 Δ(ybtA-fyuA)::FRT; Strr | This study |
| M1/5 HPI- csrA51 | M1/5 Δ(ybtA-fyuA)::FRT csrA153::FRT-kan-FRT; Kanr, Strr | This study |
| M1/5 Kan-PlacUV5-clbQ'-'venus | M1/5 ΔPclbQ::(FRT-kan-FRT-PlacUV5) ΔclbQ4-723::(venus-cat); Cmr, Kanr, Strr | This study |
| M1/5 PclbQ-AL-venus | M1/5 Δ(clbQ 5′UTR-clbQ)::(artificial 5′UTR-venus-cat); Cmr, Strr | This study |
| M1/5 PclbQ-AL-venus csrA51 | M1/5 Δ(clbQ 5′UTR-clbQ)::(artificial 5′UTR -venus-cat) csrA153:: FRT-kan-FRT; Cmr, Kanr, Strr | This study |
| M1/5 PclbQ-clbQ'-'venus | M1/5 ΔclbQ4-723::(venus-cat); pks+, HPI+, Cmr, Strr | This study |
| M1/5 PclbQ-clbQ'-'venus csrA51 | M1/5 ΔclbQ4-723::(venus-cat) csrA153:: FRT-kan-FRT; Cmr, Kanr, Strr | This study |
| M1/5 PlacUV5-clbQ'-'venus | M1/5 ΔPclbQ::(FRT-PlacUV5) ΔclbQ4-723::(venus-cat); Cmr, Strr | This study |
| M1/5 PlacUV5-clbQ'-'venus csrA51 | M1/5 ΔPclbQ::(FRT-PlacUV5) ΔclbQ4-723::(venus-cat) csrA153::FRT-kan-FRT; Cmr, Kanr, Strr | This study |
| M1/5 PlacUV5-clbQ*'-'venus | M1/5 ΔPclbQ::(FRT-PlacUV5) ΔclbQ4-723::(venus-cat); carries modified nucleotides in clbQ 5′UTR; Cmr, Strr | This study |
| M1/5 PlacUV5-clbQ*'-'venus csrA51 | M1/5 ΔPclbQ::(FRT-PlacUV5) ΔclbQ4-723::(venus-cat) csrA153::FRT-kan-FRT; carries modified nucleotides in clbQ 5′UTR; Cmr, Kanr, Strr | This study |
| M1/5 PlacUV5-ybtA'-'venus | M1/5 ΔPybtA::(FRT-PlacUV5) ΔybtA4-960::(venus-cat); Cmr, Strr | This study |
| M1/5 PybtA-ybtA'-'venus | M1/5 ΔybtA4-960::(venus-cat); Cmr, Strr | This study |
| MG1655 | K-12 F– λ– ilvG– rfb-50 rph-1; pks–, HPI– | 94 |
| SP15 | neonatal meningitis isolate, pks+, HPI+, Strr | 95 |
| SP15 ΔuvrY | SP15 ΔuvrY::cat; Cmr, Strr | This study |
| SP15 ΔcsrB | SP15 ΔcsrB::cat; Cmr, Strr | This study |
| SP15 ΔcsrC | SP15 ΔcsrC::tet; Strr, Tetr | This study |
| SP15 ΔcsrB ΔcsrC | SP15 ΔcsrB::FRT ΔcsrC::tet; Strr, Tetr | This study |
| SY327λpir | λ(lac pro) argE (Am) rif nalA recA56 (λpir) | 96 |
| Salmonella enterica Typhimurium | ||
| WR1542 | fepA::Tn10dTc, iroN::pGP704 cir::MudJ carrying plasmid pACYC5.3L; Apr, Cmr, Kanr, Tcr | Gift from W. Rabsch, Wernigerode |
TABLE 5.
Plasmids used in this study
| Plasmid | Features | Reference |
|---|---|---|
| pBAD33 | Medium copy vector; p15A araC ParaBAD; Cmr | 97 |
| pBAD33* | pBAD33; ΔaraC ΔParaBAD | This study |
| pBAD33-csrA | pBAD33* with PcsrA-csrA | This study |
| pBAD33-venus | pBAD33* with promoterless venus | 45 |
| pBAD33-venus-csrA | pBAD33* with PcsrA-csrA and promoterless venus | This study |
| pCA9505 | gal uvrYC; Apr | 50 |
| pCA9505-MluI | pCA9505 with ΔuvrY | 90 |
| pCP20 | temp-sensitive origin of replication, encodes Flp recombinase; Apr, Cmr | 98 |
| pGEM-csrC | pGEM-T with PcsrC-csrC | This study |
| pKD3 | Template plasmid for amplification of the FRT-flanked chloramphenicol resistance cassette; Apr, Cmr | 89 |
| pKD4 | Template plasmid for amplification of the FRT-flanked kanamycin resistance cassette; Apr, Kanr | 89 |
| pKD46 | Helper plasmid for l-arabinose inducible expression of λ-Red recombinase (araC ParaB-γ-β-exo); Apr | 89 |
| pKD46-csrA | pKD46 with PcsrA-csrA | This study |
| pPlacUV5-AL-venus | pBAD33* with fusion of lacUV5 promoter, artificial 5′UTR and venus | This study |
| pPybtAALvenus | pBAD33* with fusion of ybtA promoter, artificial 5′UTR and venus | This study |
| pPybtA-ybtA'-'venus2 | pBAD33* with fusion of ybtA promoter, ybtA 5′UTR and ybtA(1-12)-venus | This study |
| pPybtA-ybtA'-'venus2-csrA | pBAD33-csrA with fusion of ybtA promoter, ybtA 5′UTR and ybtA(1-12)-venus | This study |
| pPybtA-ybtA*'-'venus2 | pBAD33* with fusion of ybtA promoter, modified ybtA 5′UTR and ybtA(1-12)-venus | This study |
| pRS-csrB | pRS1553 with PcsrB-csrB | This study |
| pUC-PlacUV5-venus | pUC18 template vector for amplification of fusion lacUV5 promoter, artificial 5′UTR and venus; Apr, Cmr | This study |
| pWKS30 | Single copy vector; pSC101 origin of replication; lacZα; Apr | 99 |
| pWKS-csrA | pWKS30 with PcsrA-csrA | This study |
Unless indicated otherwise, bacteria were grown in lysogeny broth (LB) (10 g · L−1 tryptone, 5 g · L−1 yeast extract, and 5 g · L−1 sodium chloride) or in a modified and glucose-free M9 medium (88) containing sodium pyruvate and casein hydrolysate (12 g · L−1 disodium hydrogen phosphate, 3 g · L−1 potassium dihydrogen phosphate, 3 g · L−1 casein hydrolysate, 2 g · L−1 sodium pyruvate, 1 g · L−1 ammonium chloride, 0.46 g · L−1 sodium chloride, 0.24 g · L−1 magnesium sulfate, 0.011 g · L−1 calcium chloride, and 0.2 mg · L−1 thiamine hydrochloride). For solid media, agar was used in concentrations of 16 g · L−1 in LB or 20 g · L−1 in M9. If required, 100 μM 2,2′-dipyridyl was added. The following antibiotics, if needed, were applied at the indicated concentrations: ampicillin, 100 μg · mL−1; chloramphenicol, 15 μg · mL−1 and 25 μg · mL−1 for low and medium copy number of the resistance cassette, respectively; kanamycin, 50 μg · mL−1; and tetracycline, 10 μg · mL−1.
Chromosomal genetic manipulations were carried out using the lambda red recombinase system according to Datsenko and Wanner (89). A detailed description of the construction of plasmids and mutants is found in the Supplemental Material. Unless otherwise indicated, E. coli strain DH5α was used as a host for the cloning of plasmids. Oligonucleotides used for strain manipulations and construction of plasmids are given in Table S1 in the supplemental material.
Oligonucleotides used in this study. Download Table S1, PDF file, 0.2 MB (209.9KB, pdf) .
Copyright © 2022 Rehm et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Megalocytosis and γH2AX assays.
HeLa cells, maintained by serial passage in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS) and nonessential amino acids at 37°C and 5% CO2, were used to demonstrate the cytotoxic effect of colibactin on mammalian cells. Colibactin has been shown to induce double-strand breaks, which leads to cell cycle arrest and therefore the formation of megalocytotic cells (22). Assays to demonstrate megalocytosis and DNA damage induced by colibactin-producing bacteria were performed as described previously (22, 34). A detailed description is provided in the Supplemental Material.
N-Myristoyl-d-asparagine (C14-asparagine) quantification.
N-Myristoyl-d-asparagine levels were quantified in cultures grown for 24 h in 9.5 mL DMEM-HEPES (Gibco). For details, see the Supplemental Material.
DNA cross-linking assay.
The assay was performed as described previously (27). For details, please see the information provided in the Supplemental Material.
Reporter gene measurements.
Fluorescence of the yellow fluorescent protein Venus was measured to determine the expression levels of various E. coli M1/5 clbQ- and ybtA-venus fusion strains. The reporter gene assay is described in detail in the Supplemental Material.
RNA electrophoretic mobility shift assays (RNA EMSAs).
EMSAs with the purified CsrA protein and RNA molecules representing the 5′ untranslated leader regions of clbQ and ybtA were carried out using the LightShift chemiluminescent RNA EMSA kit (Thermo Fisher Scientific). For details, please see the Supplemental Material.
Yersiniabactin quantification.
The amount of yersiniabactin produced by various E. coli strains was quantified using a reporter gene-based method that has been described in detail previously (42).
Statistical analysis.
Statistical analyses were performed using the GraphPad Prism software (version 6.0). Figures show the mean values with standard deviation (STDEV.P). Either a one-way analysis of variance (ANOVA) followed by a Bonferroni posttest or Kruskal-Wallis followed by a Tukey test was applied unless stated otherwise. A P value of <0.05 was considered statistically significant, and P values are indicated by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Detailed description of cloning procedures and experimental methods. Download Text S1, PDF file, 0.4 MB (431.2KB, pdf) .
Copyright © 2022 Rehm et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
The work of the Münster team was supported by the Interdisciplinary Center for Clinical Research of the Medical Faculty Münster (Dob2/013/12) and the German Research Foundation (DO789/11-1). The work of the Würzburg team was supported by the German Research Foundation (SFB 479, TP A1). The work of the team in Toulouse was supported by research grant ANR-16-CE18-0011.
We also thank the MetaToul Lipidomics facility (INSERM UMR1048, Toulouse, France) for the quantification of metabolites. We thank K. Tegelkamp, O. Mantel, and M. van Cann (Münster) for excellent technical support. Support of H. Klimek and M. Keizers by the Research Training Group 2220 EvoPAD (281125614/GRK 2220, German Research Foundation) is gratefully acknowledged. Data reported in this study appear in part in the PhD theses of S. Homburg and A. Wallenstein.
Contributor Information
Ulrich Dobrindt, Email: dobrindt@uni-muenster.de.
Vanessa Sperandio, University of Texas Southwestern Medical Center Dallas.
REFERENCES
- 1.Putze J, Hennequin C, Nougayrède JP, Zhang W, Homburg S, Karch H, Bringer MA, Fayolle C, Carniel E, Rabsch W, Oelschlaeger TA, Oswald E, Forestier C, Hacker J, Dobrindt U. 2009. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect Immun 77:4696–4703. doi: 10.1128/IAI.00522-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wami H, Wallenstein A, Sauer D, Stoll M, von Bünau R, Oswald E, Müller R, Dobrindt U. 2021. Insights into evolution and coexistence of the colibactin- and yersiniabactin secondary metabolite determinants in enterobacterial populations. Microb Genom 7:e000577. doi: 10.1099/mgen.0.000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auvray F, Perrat A, Arimizu Y, Chagneau CV, Bossuet-Greif N, Massip C, Brugère H, Nougayrède JP, Hayashi T, Branchu P, Ogura Y, Oswald E. 2021. Insights into the acquisition of the pks island and production of colibactin in the Escherichia coli population. Microb Genom 7:e000579. doi: 10.1099/mgen.0.000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heesemann J, Hantke K, Vocke T, Saken E, Rakin A, Stojiljkovic I, Berner R. 1993. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65,000 Da and pesticin sensitivity. Mol Microbiol 8:397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 5.Karch H, Schubert S, Zhang D, Zhang W, Schmidt H, Olschläger T, Hacker J. 1999. A genomic island, termed high-pathogenicity island, is present in certain non-O157 Shiga toxin-producing Escherichia coli clonal lineages. Infect Immun 67:5994–6001. doi: 10.1128/IAI.67.11.5994-6001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rakin A, Saken E, Harmsen D, Heesemann J. 1994. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol Microbiol 13:253–263. doi: 10.1111/j.1365-2958.1994.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 7.Schubert S, Rakin A, Heesemann J. 2004. The Yersinia high-pathogenicity island (HPI): evolutionary and functional aspects. Int J Med Microbiol 294:83–94. doi: 10.1016/j.ijmm.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 8.Bobrov AG, Kirillina O, Fetherston JD, Miller MC, Burlison JA, Perry RD. 2014. The Yersinia pestis siderophore, yersiniabactin, and the ZnuABC system both contribute to zinc acquisition and the development of lethal septicaemic plague in mice. Mol Microbiol 93:759–775. doi: 10.1111/mmi.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koh EI, Hung CS, Parker KS, Crowley JR, Giblin DE, Henderson JP. 2015. Metal selectivity by the virulence-associated yersiniabactin metallophore system. Metallomics 7:1011–1022. doi: 10.1039/c4mt00341a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koh EI, Robinson AE, Bandara N, Rogers BE, Henderson JP. 2017. Copper import in Escherichia coli by the yersiniabactin metallophore system. Nat Chem Biol 13:1016–1021. doi: 10.1038/nchembio.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson AE, Lowe JE, Koh EI, Henderson JP. 2018. Uropathogenic enterobacteria use the yersiniabactin metallophore system to acquire nickel. J Biol Chem 293:14953–14961. doi: 10.1074/jbc.RA118.004483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brumbaugh AR, Smith SN, Subashchandrabose S, Himpsl SD, Hazen TH, Rasko DA, Mobley HL. 2015. Blocking yersiniabactin import attenuates extraintestinal pathogenic Escherichia coli in cystitis and pyelonephritis and represents a novel target to prevent urinary tract infection. Infect Immun 83:1443–1450. doi: 10.1128/IAI.02904-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaturvedi KS, Hung CS, Crowley JR, Stapleton AE, Henderson JP. 2012. The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat Chem Biol 8:731–736. doi: 10.1038/nchembio.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson JR, Russo TA. 2018. Molecular epidemiology of extraintestinal pathogenic Escherichia coli. EcoSal Plus 8:2324-6200. doi: 10.1128/ecosalplus.ESP-0004-2017. [DOI] [PubMed] [Google Scholar]
- 15.Magistro G, Magistro C, Stief CG, Schubert S. 2017. The high-pathogenicity island (HPI) promotes flagellum-mediated motility in extraintestinal pathogenic Escherichia coli. PLoS One 12:e0183950. doi: 10.1371/journal.pone.0183950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schubert S, Picard B, Gouriou S, Heesemann J, Denamur E. 2002. Yersinia high-pathogenicity island contributes to virulence in Escherichia coli causing extraintestinal infections. Infect Immun 70:5335–5337. doi: 10.1128/IAI.70.9.5335-5337.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. 1998. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect Immun 66:480–485. doi: 10.1128/IAI.66.2.480-485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smati M, Magistro G, Adiba S, Wieser A, Picard B, Schubert S, Denamur E. 2017. Strain-specific impact of the high-pathogenicity island on virulence in extra-intestinal pathogenic Escherichia coli. Int J Med Microbiol 307:44–56. doi: 10.1016/j.ijmm.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Su Q, Guan T, Lv H. 2016. Siderophore biosynthesis coordinately modulated the virulence-associated interactive metabolome of uropathogenic Escherichia coli and human urine. Sci Rep 6:24099. doi: 10.1038/srep24099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anisimov R, Brem D, Heesemann J, Rakin A. 2005. Transcriptional regulation of high pathogenicity island iron uptake genes by YbtA. Int J Med Microbiol 295:19–28. doi: 10.1016/j.ijmm.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Anisimov R, Brem D, Heesemann J, Rakin A. 2005. Molecular mechanism of YbtA-mediated transcriptional regulation of divergent overlapping promoters ybtA and irp6 of Yersinia enterocolitica. FEMS Microbiol Lett 250:27–32. doi: 10.1016/j.femsle.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 22.Nougayrède JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. 2006. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 23.Li ZR, Li J, Cai W, Lai JYH, McKinnie SMK, Zhang WP, Moore BS, Zhang W, Qian PY. 2019. Macrocyclic colibactin induces DNA double-strand breaks via copper-mediated oxidative cleavage. Nat Chem 11:880–889. doi: 10.1038/s41557-019-0317-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue M, Kim CS, Healy AR, Wernke KM, Wang Z, Frischling MC, Shine EE, Wang W, Herzon SB, Crawford JM. 2019. Structure elucidation of colibactin and its DNA cross-links. Science 365:eaax2685. doi: 10.1126/science.aax2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang Y, Stornetta A, Villalta PW, Wilson MR, Boudreau PD, Zha L, Balbo S, Balskus EP. 2019. Reactivity of an unusual amidase may explain colibactin’s DNA cross-linking activity. J Am Chem Soc 141:11489–11496. doi: 10.1021/jacs.9b02453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson MR, Jiang Y, Villalta PW, Stornetta A, Boudreau PD, Carrá A, Brennan CA, Chun E, Ngo L, Samson LD, Engelward BP, Garrett WS, Balbo S, Balskus EP. 2019. The human gut bacterial genotoxin colibactin alkylates DNA. Science 363:eaar7785. doi: 10.1126/science.aar7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bossuet-Greif N, Vignard J, Taieb F, Mirey G, Dubois D, Petit C, Oswald E, Nougayrède JP. 2018. The colibactin genotoxin generates DNA interstrand cross-links in infected cells. mBio 9:e02393-17. doi: 10.1128/mBio.02393-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marcq I, Martin P, Payros D, Cuevas-Ramos G, Boury M, Watrin C, Nougayrède JP, Olier M, Oswald E. 2014. The genotoxin colibactin exacerbates lymphopenia and decreases survival rate in mice infected with septicemic Escherichia coli. J Infect Dis 210:285–294. doi: 10.1093/infdis/jiu071. [DOI] [PubMed] [Google Scholar]
- 29.McCarthy AJ, Martin P, Cloup E, Stabler RA, Oswald E, Taylor PW. 2015. The genotoxin colibactin is a determinant of virulence in Escherichia coli K1 experimental neonatal systemic infection. Infect Immun 83:3704–3711. doi: 10.1128/IAI.00716-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olier M, Marcq I, Salvador-Cartier C, Secher T, Dobrindt U, Boury M, Bacquié V, Pénary M, Gaultier E, Nougayrède JP, Fioramonti J, Oswald E. 2012. Genotoxicity of Escherichia coli Nissle 1917 strain cannot be dissociated from its probiotic activity. Gut Microbes 3:501–509. doi: 10.4161/gmic.21737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pérez-Berezo T, Pujo J, Martin P, Le Faouder P, Galano JM, Guy A, Knauf C, Tabet JC, Tronnet S, Barreau F, Heuillet M, Dietrich G, Bertrand-Michel J, Durand T, Oswald E, Cenac N. 2017. Identification of an analgesic lipopeptide produced by the probiotic Escherichia coli strain Nissle 1917. Nature Commun 8:1314. doi: 10.1038/s41467-017-01403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vizcaino MI, Engel P, Trautman E, Crawford JM. 2014. Comparative metabolomics and structural characterizations illuminate colibactin pathway-dependent small molecules. J Am Chem Soc 136:9244–9247. doi: 10.1021/ja503450q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, Déchelotte P, Bonnet R, Pezet D, Darfeuille-Michaud A. 2014. Colonization of the human gut by E. coli and colorectal cancer risk. Clin Cancer Res 20:859–867. doi: 10.1158/1078-0432.CCR-13-1343. [DOI] [PubMed] [Google Scholar]
- 34.Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède JP. 2010. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci USA 107:11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dziubańska-Kusibab PJ, Berger H, Battistini F, Bouwman BAM, Iftekhar A, Katainen R, Cajuso T, Crosetto N, Orozco M, Aaltonen LA, Meyer TF. 2020. Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat Med 26:1063–1069. doi: 10.1038/s41591-020-0908-2. [DOI] [PubMed] [Google Scholar]
- 36.Iftekhar A, Berger H, Bouznad N, Heuberger J, Boccellato F, Dobrindt U, Hermeking H, Sigal M, Meyer TF. 2021. Genomic aberrations after short-term exposure to colibactin-producing E. coli transform primary colon epithelial cells. Nat Commun 12:1003. doi: 10.1038/s41467-021-21162-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, Gurjao C, Manders F, Dalmasso G, Stege PB, Paganelli FL, Geurts MH, Beumer J, Mizutani T, Miao Y, van der Linden R, van der Elst S, Garcia KC, Top J, Willems RJL, Giannakis M, Bonnet R, Quirke P, Meyerson M, Cuppen E, van Boxtel R, Clevers H, Genomics England Research Consortium . 2020. Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature 580:269–273. doi: 10.1038/s41586-020-2080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chagneau CV, Garcie C, Bossuet-Greif N, Tronnet S, Brachmann AO, Piel J, Nougayrède JP, Martin P, Oswald E. 2019. The polyamine spermidine modulates the production of the bacterial genotoxin colibactin. mSphere 4:e00414-19. doi: 10.1128/mSphere.00414-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang-Fichaux M, Chagneau CV, Bossuet-Greif N, Nougayrède JP, Oswald E, Branchu P. 2020. The polyphosphate kinase of Escherichia coli is required for full production of the genotoxin colibactin. mSphere 5:e01195-20. doi: 10.1128/mSphere.01195-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wallenstein A, Rehm N, Brinkmann M, Selle M, Bossuet-Greif N, Sauer D, Bunk B, Sproer C, Wami HT, Homburg S, von Bünau R, König S, Nougayrède JP, Overmann J, Oswald E, Müller R, Dobrindt U. 2020. ClbR is the key transcriptional activator of colibactin gene expression in Escherichia coli. mSphere 5:e00591-20. doi: 10.1128/mSphere.00591-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliero M, Calvé A, Fragoso G, Cuisiniere T, Hajjar R, Dobrindt U, Santos MM. 2021. Oligosaccharides increase the genotoxic effect of colibactin produced by pks+ Escherichia coli strains. BMC Cancer 21:172. doi: 10.1186/s12885-021-07876-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin P, Marcq I, Magistro G, Penary M, Garcie C, Payros D, Boury M, Olier M, Nougayrède JP, Audebert M, Chalut C, Schubert S, Oswald E. 2013. Interplay between siderophores and colibactin genotoxin biosynthetic pathways in Escherichia coli. PLoS Pathog 9:e1003437. doi: 10.1371/journal.ppat.1003437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin P, Tronnet S, Garcie C, Oswald E. 2017. Interplay between siderophores and colibactin genotoxin in Escherichia coli. IUBMB Life 69:435–441. doi: 10.1002/iub.1612. [DOI] [PubMed] [Google Scholar]
- 44.Tronnet S, Garcie C, Brachmann AO, Piel J, Oswald E, Martin P. 2017. High iron supply inhibits the synthesis of the genotoxin colibactin by pathogenic Escherichia coli through a non-canonical Fur/RyhB-mediated pathway. Pathog Dis 75:ftx066. doi: 10.1093/femspd/ftx066. [DOI] [PubMed] [Google Scholar]
- 45.Tronnet S, Garcie C, Rehm N, Dobrindt U, Oswald E, Martin P. 2016. Iron homeostasis regulates the genotoxicity of Escherichia coli that produces colibactin. Infect Immun 84:3358–3368. doi: 10.1128/IAI.00659-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcie C, Tronnet S, Garénaux A, McCarthy AJ, Brachmann AO, Pénary M, Houle S, Nougayrède JP, Piel J, Taylor PW, Dozois CM, Genevaux P, Oswald E, Martin P. 2016. The bacterial stress-responsive Hsp90 chaperone (HtpG) is required for the production of the genotoxin colibactin and the siderophore yersiniabactin in Escherichia coli. J Infect Dis 214:916–924. doi: 10.1093/infdis/jiw294. [DOI] [PubMed] [Google Scholar]
- 47.Berger H, Hacker J, Juarez A, Hughes C, Goebel W. 1982. Cloning of the chromosomal determinants encoding hemolysin production and mannose-resistant hemagglutination in Escherichia coli. J Bacteriol 152:1241–1247. doi: 10.1128/jb.152.3.1241-1247.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagy G, Altenhoefer A, Knapp O, Maier E, Dobrindt U, Blum-Oehler G, Benz R, Emödy L, Hacker J. 2006. Both alpha-haemolysin determinants contribute to full virulence of uropathogenic Escherichia coli strain 536. Microbes Infect 8:2006–2012. doi: 10.1016/j.micinf.2006.02.029. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki K, Wang X, Weilbacher T, Pernestig AK, Melefors O, Georgellis D, Babitzke P, Romeo T. 2002. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J Bacteriol 184:5130–5140. doi: 10.1128/JB.184.18.5130-5140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Sluis CA, Moolenaar GF, Backendorf C. 1983. Regulation of the uvrC gene of Escherichia coli K12: localization and characterization of a damage-inducible promoter. EMBO J 2:2313–2318. doi: 10.1002/j.1460-2075.1983.tb01739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romeo T, Vakulskas CA, Babitzke P. 2013. Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ Microbiol 15:313–324. doi: 10.1111/j.1462-2920.2012.02794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pannuri A, Vakulskas CA, Zere T, McGibbon LC, Edwards AN, Georgellis D, Babitzke P, Romeo T. 2016. Circuitry linking the catabolite repression and Csr global regulatory systems of Escherichia coli. J Bacteriol 198:3000–3015. doi: 10.1128/JB.00454-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weilbacher T, Suzuki K, Dubey AK, Wang X, Gudapaty S, Morozov I, Baker CS, Georgellis D, Babitzke P, Romeo T. 2003. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol 48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- 54.Romeo T, Gong M, Liu MY, Brun-Zinkernagel AM. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J Bacteriol 175:4744–4755. doi: 10.1128/jb.175.15.4744-4755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dubey AK, Baker CS, Romeo T, Babitzke P. 2005. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA 11:1579–1587. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romeo T, Babitzke P. 2018. Global regulation by CsrA and its RNA antagonists. Microbiol Spectr 6:6.2.05. doi: 10.1128/microbiolspec.RWR-0009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jonas K, Edwards AN, Ahmad I, Romeo T, Romling U, Melefors O. 2010. Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella Typhimurium. Environ Microbiol 12:524–540. doi: 10.1111/j.1462-2920.2009.02097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schubert S, Darlu P, Clermont O, Wieser A, Magistro G, Hoffmann C, Weinert K, Tenaillon O, Matic I, Denamur E. 2009. Role of intraspecies recombination in the spread of pathogenicity islands within the Escherichia coli species. PLoS Pathog 5:e1000257. doi: 10.1371/journal.ppat.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Messerer M, Fischer W, Schubert S. 2017. Investigation of horizontal gene transfer of pathogenicity islands in Escherichia coli using next-generation sequencing. PLoS One 12:e0179880. doi: 10.1371/journal.pone.0179880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Staggs TM, Perry RD. 1995. Fur regulation in Yersinia species. Mol Microbiol 17:601. [PubMed] [Google Scholar]
- 61.Massé E, Gottesman S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA 99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salvail H, Lanthier-Bourbonnais P, Sobota JM, Caza M, Benjamin JA, Mendieta ME, Lepine F, Dozois CM, Imlay J, Massé E. 2010. A small RNA promotes siderophore production through transcriptional and metabolic remodeling. Proc Natl Acad Sci USA 107:15223–15228. doi: 10.1073/pnas.1007805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leistra AN, Gelderman G, Sowa SW, Moon-Walker A, Salis HM, Contreras LM. 2018. A canonical biophysical model of the CsrA global regulator suggests flexible regulator-target interactions. Sci Rep 8:9892. doi: 10.1038/s41598-018-27474-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fetherston JD, Bearden SW, Perry RD. 1996. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol Microbiol 22:315–325. doi: 10.1046/j.1365-2958.1996.00118.x. [DOI] [PubMed] [Google Scholar]
- 65.Gao H, Zhou D, Li Y, Guo Z, Han Y, Song Y, Zhai J, Du Z, Wang X, Lu J, Yang R. 2008. The iron-responsive Fur regulon in Yersinia pestis. J Bacteriol 190:3063–3075. doi: 10.1128/JB.01910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Staggs TM, Fetherston JD, Perry RD. 1994. Pleiotropic effects of a Yersinia pestis fur mutation. J Bacteriol 176:7614–7624. doi: 10.1128/jb.176.24.7614-7624.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heroven AK, Nuss AM, Dersch P. 2017. RNA-based mechanisms of virulence control in Enterobacteriaceae. RNA Biol 14:471–487. doi: 10.1080/15476286.2016.1201617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holmqvist E, Wright PR, Li L, Bischler T, Barquist L, Reinhardt R, Backofen R, Vogel J. 2016. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J 35:991–1011. doi: 10.15252/embj.201593360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oliva G, Sahr T, Buchrieser C. 2015. Small RNAs, 5' UTR elements and RNA-binding proteins in intracellular bacteria: impact on metabolism and virulence. FEMS Microbiol Rev 39:331–349. doi: 10.1093/femsre/fuv022. [DOI] [PubMed] [Google Scholar]
- 70.Vakulskas CA, Potts AH, Babitzke P, Ahmer BM, Romeo T. 2015. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol Mol Biol Rev 79:193–224. doi: 10.1128/MMBR.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Assche E, van Puyvelde S, Vanderleyden J, Steenackers HP. 2015. RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front Microbiol 6:141. doi: 10.3389/fmicb.2015.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brencic A, Lory S. 2009. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol 72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Burrowes E, Baysse C, Adams C, O'Gara F. 2006. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology (Reading) 152:405–418. doi: 10.1099/mic.0.28324-0. [DOI] [PubMed] [Google Scholar]
- 74.Heroven AK, Bohme K, Dersch P. 2012. The Csr/Rsm system of Yersinia and related pathogens: a post-transcriptional strategy for managing virulence. RNA Biol 9:379–391. doi: 10.4161/rna.19333. [DOI] [PubMed] [Google Scholar]
- 75.Heroven AK, Bohme K, Rohde M, Dersch P. 2008. A Csr-type regulatory system, including small non-coding RNAs, regulates the global virulence regulator RovA of Yersinia pseudotuberculosis through RovM. Mol Microbiol 68:1179–1195. doi: 10.1111/j.1365-2958.2008.06218.x. [DOI] [PubMed] [Google Scholar]
- 76.Lawhon SD, Frye JG, Suyemoto M, Porwollik S, McClelland M, Altier C. 2003. Global regulation by CsrA in Salmonella Typhimurium. Mol Microbiol 48:1633–1645. doi: 10.1046/j.1365-2958.2003.03535.x. [DOI] [PubMed] [Google Scholar]
- 77.Sahr T, Rusniok C, Impens F, Oliva G, Sismeiro O, Coppee JY, Buchrieser C. 2017. The Legionella pneumophila genome evolved to accommodate multiple regulatory mechanisms controlled by the CsrA-system. PLoS Genet 13:e1006629. doi: 10.1371/journal.pgen.1006629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tan Y, Liu ZY, Liu Z, Zheng HJ, Li FL. 2015. Comparative transcriptome analysis between csrA-disruption Clostridium acetobutylicum and its parent strain. Mol Biosyst 11:1434–1442. doi: 10.1039/c4mb00600c. [DOI] [PubMed] [Google Scholar]
- 79.Berndt V, Beckstette M, Volk M, Dersch P, Brönstrup M. 2019. Metabolome and transcriptome-wide effects of the carbon storage regulator A in enteropathogenic Escherichia coli. Sci Rep 9:138. doi: 10.1038/s41598-018-36932-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Duffy BK, Defago G. 2000. Controlling instability in gacS-gacA regulatory genes during inoculant production of Pseudomonas fluorescens biocontrol strains. Appl Environ Microbiol 66:3142–3150. doi: 10.1128/AEM.66.8.3142-3150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Whistler CA, Ruby EG. 2003. GacA regulates symbiotic colonization traits of Vibrio fischeri and facilitates a beneficial association with an animal host. J Bacteriol 185:7202–7212. doi: 10.1128/JB.185.24.7202-7212.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krin E, Derzelle S, Bedard K, Adib-Conquy M, Turlin E, Lenormand P, Hullo MF, Bonne I, Chakroun N, Lacroix C, Danchin A. 2008. Regulatory role of UvrY in adaptation of Photorhabdus luminescens growth inside the insect. Environ Microbiol 10:1118–1134. doi: 10.1111/j.1462-2920.2007.01528.x. [DOI] [PubMed] [Google Scholar]
- 83.Wolfe CJ, Kohane IS, Butte AJ. 2005. Systematic survey reveals general applicability of “guilt-by-association” within gene coexpression networks. BMC Bioinformatics 6:227. doi: 10.1186/1471-2105-6-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krieger JN, Dobrindt U, Riley DE, Oswald E. 2011. Acute Escherichia coli prostatitis in previously health young men: bacterial virulence factors, antimicrobial resistance, and clinical outcomes. Urology 77:1420–1425. doi: 10.1016/j.urology.2010.12.059. [DOI] [PubMed] [Google Scholar]
- 85.Nowrouzian FL, Oswald E. 2012. Escherichia coli strains with the capacity for long-term persistence in the bowel microbiota carry the potentially genotoxic pks island. Microb Pathog 53:180–182. doi: 10.1016/j.micpath.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 86.Wassenaar TM. 2018. E. coli and colorectal cancer: a complex relationship that deserves a critical mindset. Crit Rev Microbiol 44:619–632. doi: 10.1080/1040841X.2018.1481013. [DOI] [PubMed] [Google Scholar]
- 87.Hoffmeister D, Keller NP. 2007. Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep 24:393–416. doi: 10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- 88.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 89.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pernestig AK, Georgellis D, Romeo T, Suzuki K, Tomenius H, Normark S, Melefors O. 2003. The Escherichia coli BarA-UvrY two-component system is needed for efficient switching between glycolytic and gluconeogenic carbon sources. J Bacteriol 185:843–853. doi: 10.1128/JB.185.3.843-853.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 92.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 93.Achtman M, Mercer A, Kusecek B, Pohl A, Heuzenroeder M, Aaronson W, Sutton A, Silver RP. 1983. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect Immun 39:315–335. doi: 10.1128/iai.39.1.315-335.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 95.Johnson JR, Oswald E, O'Bryan TT, Kuskowski MA, Spanjaard L. 2002. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in the Netherlands. J Infect Dis 185:774–784. doi: 10.1086/339343. [DOI] [PubMed] [Google Scholar]
- 96.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 99.Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of the genomic region comprising uvrY in different E. coli strains. Analysis of the chromosomal region next to uvrY in the complete genome sequences of pks island-positive, but CPE-negative, uropathogenic E. coli strain 536 with those of colibactin-producing newborn meningitis isolate IHE3034 as well as to K-12 laboratory strain MG1655 that indicated that E. coli 536 carries a 6.1-kb chromosomal deletion spanning the region between uvrC and fliY and thus lacks the uvrY gene. Download FIG S1, PDF file, 0.2 MB (215.3KB, pdf) .
Copyright © 2022 Rehm et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Impact of the small regulatory RNAs csrB and csrC on the cytopathic effect of E. coli strain SP15 in HeLa cells. HeLa cells were either not infected or infected with the indicated E. coli strains to a multiplicity of infection (MOI) of 250. After 4 hours of infection, HeLa cells were washed to remove bacteria and further cultivated. At 72 h postinfection, cells were washed and Giemsa stained. Scale bars, 100 μm. Plasmid pRS-csrB was used to complement the csrB and csrC deletion in E. coli strain SP15ΔcsrB ΔcsrC. Download FIG S2, PDF file, 0.4 MB (382.3KB, pdf) .
Copyright © 2022 Rehm et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Activity of the clbQ promoter fused to a sequence encoding an artificial 5′ untranslated leader and venus as a reporter gene. (A) Sequences encoding the clbQ 5′ leader and the artificial leader are shown in italics, an asterisk represents the transcription start, and the translational start codon is depicted in bold. The sequence encoding the putative CsrA-binding motif is underlined. (B) Expression from the clbQ promoter in E. coli strains M1/5 PclbQ-AL-'venus and M1/5 PclbQ-AL-'venus was measured by means of venus fluorescence after 10 h of growth in M9 minimal medium without glucose but containing pyruvate and casein hydrolysate. Mean values with standard deviations of three independent experiments are shown. Download FIG S3, PDF file, 0.2 MB (248.3KB, pdf) .
Copyright © 2022 Rehm et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotides used in this study. Download Table S1, PDF file, 0.2 MB (209.9KB, pdf) .
Copyright © 2022 Rehm et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detailed description of cloning procedures and experimental methods. Download Text S1, PDF file, 0.4 MB (431.2KB, pdf) .
Copyright © 2022 Rehm et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.