Abstract
Background
The novel beta-coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), enters the human body via mucosal surfaces of the upper and/or lower respiratory tract. Viral entry into epithelial cells is mediated via angiotensin-converting enzyme 2 (ACE2) and auxiliary molecules, but the precise anatomic site of infection still remains unclear.
Methods
Here, we systematically investigated the main SARS-CoV-2 receptor proteins ACE2 and transmembrane serine protease 2 (TMPRSS2), as well as 2 molecules potentially involved in viral entry, furin and CD147, in formalin-fixed, paraffin-embedded human tissues. Tissue microarrays incorporating a total of 879 tissue cores from conjunctival (n = 84), sinonasal (n = 95), and lung (bronchiolar/alveolar; n = 96) specimens were investigated for protein expression by immunohistochemistry.
Results
ACE2 and TMPRSS2 were expressed in ciliated epithelial cells of the conjunctivae and sinonasal tissues, with highest expression levels observed in the apical cilia. In contrast, in the lung, the expression of those molecules in bronchiolar and alveolar epithelial cells was much rarer and only very focal when present. Furin and CD147 were more uniformly expressed in all tissues analyzed, including the lung. Interestingly, alveolar macrophages consistently expressed high levels of all 4 molecules investigated.
Conclusions
Our study confirms and extends previous findings and contributes to a better understanding of potential SARS-CoV-2 infection sites along the human respiratory tract.
Keywords: Severe acute respiratory syndrome coronavirus 2, Angiotensin-converting enzyme 2, Transmembrane serine protease 2, Furin, CD147, Immunohistochemistry, Tissue microarray, Coronavirus disease-19
Introduction
Coronavirus disease-19 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which has spread all over the globe with enormous consequences on economies and health systems. Major challenges are the unpredictable nature of the viral infectious cycle and the transmission via asymptomatic carriers [1]. By late 2021, SARS-CoV-2 had infected more than 250 million people worldwide, caused over 5 million deaths, and resulted in the necessity for hospitalization and intensive care in many more [2]. SARS-CoV-2 infection may be asymptomatic or lead to a range of symptoms from a mild influenza-like illness to life-threatening acute respiratory distress syndrome with fatal outcomes in humans [3]. The detailed pathogenesis and disease biology of COVID-19 are still under investigation [4]. Local cytokine release, e.g., elevated interleukin-6 levels, has been described and is associated with a more critical clinical presentation of COVID-19 [5]. While pulmonary edema in COVID-19 is linked to the activation of the kinin-kallikrein system [6], the frequently reported thromboses are likely triggered by complement system-induced overactivation of the coagulation cascade [7].
SARS-CoV-2, a beta corona and single-stranded RNA virus, is transmitted in humans via droplets and aerosols, mainly in close contact and indoors [3]. Structurally, the virus is composed of a core and envelope proteins. Specific spike proteins of the envelope − each having S1 and S2 subunits [8] − constitute its surface. The number of spike proteins per virus is variable, and these are subject to frequent mutations. Viral spike protein plays an important role in cellular attachment and entry, important steps in the viral life cycle [4]. Notwithstanding, mechanisms of virus entry, transmission, as well as long-time sequelae after infection are insufficiently explained and are an area of active research [9].
First described in 2004 after the SARS-CoV-1 pandemic, the angiotensin-converting enzyme 2 (ACE2) receptor has been discussed as the main receptor for viral entry of Coronaviridae [9, 10, 11, 12]. Physiologically, ACE2 catalyzes the conversion of angiotensin I into angiotensin II and is thereby involved in the regulation of blood pressure via the renin-angiotensin-aldosterone system [13]. The ACE2 receptor is expressed in multiple organs and is discussed as a potential therapeutic target to prevent virus entry [14].
Recently, conflicting data on ACE2 protein expression in cells of the respiratory tract have been reported, and important controversies remain [15, 16, 17, 18, 19]. Lee et al. [15] and Nakayama et al. [16] showed homogeneous expression specifically in the cilia in the majority of ciliated respiratory cells, predominantly in the upper respiratory tract, but also in lung tissues. In contrast, Ortiz et al. [17] found ACE2 to be expressed in the ciliary apparatus only in rare epithelial cells of the trachea and larger bronchi, bronchioli, and alveoli, although in the majority of samples studied. On the other hand, Aguiar et al. [18] detected ACE2 expression in only 1 out of 98 investigated lung samples, which is in line with Hikmet et al. [19], who reported a positive rate of 0.56% using 2 different antibodies in a large cohort of 360 lung samples.
Transmembrane serine protease 2 (TMPRSS2) cleaves viral spike protein and thereby serves as an ACE2 cofactor, facilitating viral entry [20, 21]. Its inhibition is discussed as a potential therapeutic strategy [21]. However, camostat mesylate, a TMPRSS2 inhibitor, could so far not demonstrate clinical efficacy in a randomized controlled trial [22, 23]. Moreover, furin, a proprotein convertase, has been proposed as an essential activating cofactor for virus entry as well [24]. In contrast, the role of CD147 (basigin) as a viral co-receptor remains controversial [25, 26, 27].
In this study, we systematically investigated the distribution of ACE2, TMPRSS2, furin, and CD147 along viral entry routes in humans. ACE2 and TMPRSS2 expression levels were reproducibly detectable in ciliated epithelial cells of the conjunctivae and sinonasal tissues, whereas in the bronchioli and alveoli, expression was rare and focal. In contrast, furin and CD147 were more uniformly expressed, including in lung tissues. Finally, alveolar macrophages consistently expressed high levels of all 4 molecules investigated. Our study confirms and extends previous findings and contributes to a better understanding of potential sites of SARS-CoV-2 infection along the human respiratory tract.
Methods
Patient Samples
Formalin-fixed, paraffin-embedded surgical specimens of the conjunctivae, sinonasal tract, and lungs (bronchi and alveoli) were used to construct tissue microarrays (TMAs) and determine the protein expression levels of ACE2, TMPRSS2, furin, and CD147 by immunohistochemistry (IHC). Formalin-fixed, paraffin-embedded tissue blocks from 2011 to 2014 were retrieved from the archives of the Institute of Pathology, University of Bern, Switzerland. Details on the included samples are summarized in Table 1. The study was approved by the local Ethics Committee of the Canton of Bern, Switzerland (KEK 200/2014).
Table 1.
Sample characteristics
| Site | Samples | Sex, % | Age range (median), year | Prior history of malignancy, n (%) | Histopathological diagnoses, n/n (%) |
|---|---|---|---|---|---|
| Conjunctivae | 84 | 39.3 F 60.7 M |
6–93 (53) | 2 (2.4) | 50/84 pterygium (59.5) 8/84 nevus (9.5) 7/84 inflammation (8.3) 6/84 fibroepithelial polyp (7.1) 13/84 others (15.5) |
|
| |||||
| Sinonasal tract | 95 | 36.8 F 63.2 M |
0–101 (52) | 14 (14.7) | 59/95 rhinosinusitis (62.8) 21/95 maxillary sinus (22.1) 21/95 ethmoid sinus (22.1) 23/95 nasal polyp (24.5) 20/82 high eosinophil count (24.4) 16/82 low eosinophil count (19.5) 6/82 no eosinophils (7.3) 40/42 n/a (48.8) 13/95 others (13.7) |
|
| |||||
| Lung, containing both bronchioli and alveoli | 96 | 44.8 F 55.2 M |
10–85 (61) | 23 (24.0) | 50/96 malignant tumor (52.1) 29/96 adenocarcinoma (30.2) 9/96 squamous cell carcinoma (9.4) 12/96 others (metastasis, etc.) (12.5) 46/96 others (e.g., pneumonia, reactive, benign neoplasms) (47.9) 21/96 with emphysematous changes (21.9) |
Next-Generation TMA Construction and IHC
Regions of interest were annotated in CaseViewer software (3DHISTECH, Budapest, Hungary) on digitized hematoxylin and eosin-stained sections. For each specimen, 3 representative tissue cores of 0.6 mm diameter were assembled into next-generation TMAs using a Grand Master automated TMA (3DHISTECH). Blocks were cut at 2.5 μm, deparaffinized, and rehydrated in dewax solution (Leica Biosystems, Wetzlar, Germany). IHC was performed on a BOND-RX automated immunostainer (Leica Biosystems).
Primary antibodies were incubated for 30 min at room temperature and used as follows: ACE2 (MAB933, mouse monoclonal IgG2a, clone 171606; Novus Biologicals, Littleton, CO, USA); dilution: 1:24.000; retrieval: citrate buffer, 30 min, 100°C; TMPRSS2 (HPA035787, rabbit polyclonal; Sigma-Aldrich, St. Louis, MO, USA); dilution: 1:800; retrieval: Tris buffer, 20 min, 95°C; Furin (NBP2-12321, mouse monoclonal IgG1, clone MM0298-4G31; Novus Biologicals); dilution: 1:50; retrieval: Tris buffer, 30 min, 95°C; CD147 (ab666, mouse monoclonal IgG1, clone MEM-M6/1; Abcam, Cambridge, UK); dilution: 1:2,000; retrieval: Tris buffer, 30 min 95°C. Antibody detection was performed with the BOND Polymer Refine Detection kit (DS9800; Leica Biosystems) using 3,3-diaminobenzidine as the brown chromogen. The samples were counterstained with hematoxylin, dehydrated and mounted with Pertex (Sakura, Alphen aan den Rijn, the Netherlands). Slides were scanned on a Pannoramic 250 Flash scanner (3DHISTECH). Placental, testicular, prostatic, and renal tissues were used as positive and negative controls.
TMA Interpretation and Statistics
Individual TMA cores were evaluated in a blinded, independent, and randomized manner by 4 pathologists (K.B., M.W., E.H., and C.M.S.) using Scorenado, a custom-made browser-based online TMA analysis tool [28, 29]. Staining intensities were quantified in a 3-tiered manner: 0, negative; 1, low; and 2, moderate to strong expression (online suppl. material S1, S2; for all online suppl. material, see www.karger.com/doi/10.1159/000521317). Median values across all observers were used for final analyses. Decimal numbers were rounded up. Scores were only included for further analyses if tissue quality was deemed sufficient by at least 2 pathologists. Discrepant cases were jointly reviewed by K.B. and C.M.S., and a final consensus score was applied. Therefore, up to 59/84 conjunctivae (70.2%), 64/95 sinonasal samples (67.4%), 68/96 bronchioli (70.8%), and 75/96 alveoli (78.1%) were included.
Statistical analyses were performed using SPSS 26.0 (SPSS Inc., Chicago, IL, USA). The χ2 test was used to calculate contingency tables. p values <0.05 were considered statistically significant. Correlation analyses were conducted using Spearman-rho coefficients (ρ). Fleiss kappa (κ) was used to assess the strength of interobserver agreement for multiraters [30]. Interobserver agreement was “moderate” (global κ = 0.495; 95% confidence interval 0.470–0.520), calculated using the DescTools package [31] in R Studio (R Foundation for Statistical Computing, Vienna, Austria). Moreover, expression of molecules was compared in 3 arbitrarily defined age subgroups (0–30 years, 31–60 years, and >60 years).
Results
ACE2 and TMPRSS2 Protein Expression Is Detectable but Low in Conjunctival and Sinonasal Specimens
The conjunctivae and sinonasal tracts are mucosal sites that can serve as viral entry routes for SARS-CoV-2 [32, 33]. We therefore analyzed the expression of ACE2, the main receptor for viral spike protein, and its cofactor TMPRSS2, in 252 and 285 TMA cores of conjunctival and sinonasal specimens, respectively, from patients undergoing surgery for common conditions such as pterygium and chronic rhinosinusitis.
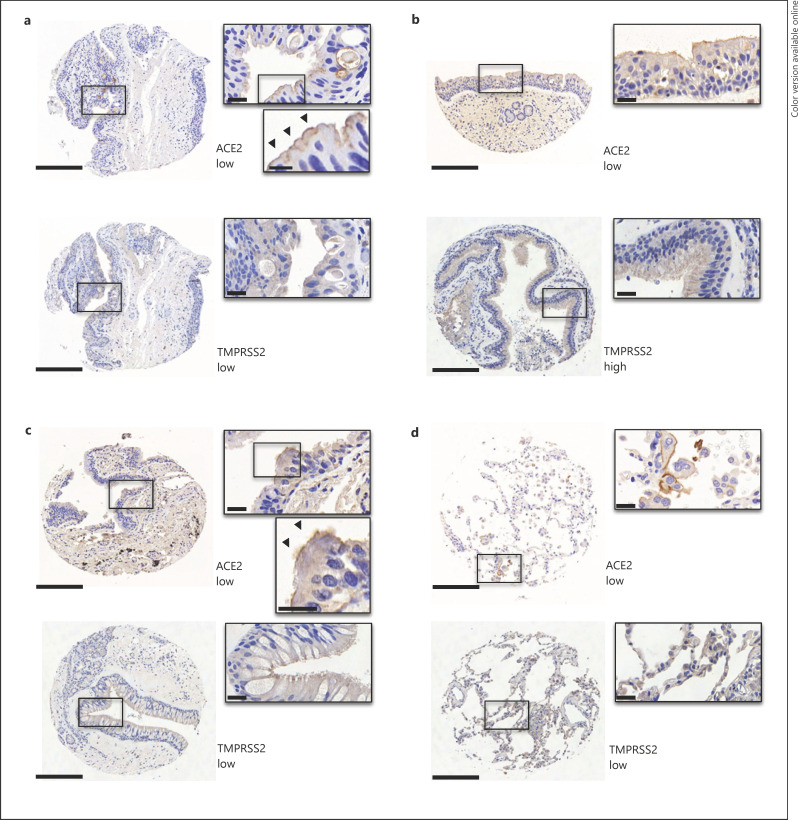
We found that 23/57 (40.4%) of conjunctival specimens showed low ACE2 expression, primarily localized to the cilia of apical respiratory epithelial cells (Fig. 1a; Table 2 and online suppl. material S1, S2). For TMPRSS2, 12/55 (21.9%) specimens had detectable low-level protein expression (Fig. 1a; Table 2); 8 of those (66.7%) also showed co-expression with ACE2.
Fig. 1.
Expression of ACE2 and TMPRSS2 in conjunctival, sinonasal, and lung tissues. Representative IHC images out of 252 and 285 TMA cores from conjunctival (a) and sinonasal specimens (b), respectively. The conjunctival specimen depicted shows metaplastic ciliated epithelium due to chronic irritation (diagnosis of pterygium). Representative IHC images out of 342 TMA cores from bronchioli (c) and alveoli (d). Scale bar, 200 μm;inset, 20 μm.
Table 2.
Receptor protein expression in conjunctivae and the sinonasal tract (%)
| Median | Negative, n/n (%) | Low, n/n (%) | High, n/n (%) | |
|---|---|---|---|---|
| Conjunctivae | ||||
| ACE2 | 0 | 34/57 (59.6) | 23/57 (40.4) | 0/57 |
| TMPRSS2 | 0 | 43/55 (78.2) | 12/55 (21.8) | 0/55 |
| Furin | 1 | 6/54 (11.1) | 43/54 (79.6) | 5/54 (9.3) |
| CD147 | 2 | 0/59 | 12/59 (20.3) | 47/59 (79.7) |
|
| ||||
| Sinonasal tract | ||||
| ACE2 | 1 | 18/64 (28.1) | 46/64 (71.9) | 0/64 |
| TMPRSS2 | 1 | 6/49 (12.2) | 39/49 (79.6) | 4/49 (8.2) |
| Furin | 1 | 5/52 (9.6) | 45/52 (86.5) | 2/52 (3.8) |
| CD147 | 2 | 0/54 | 2/54 (3.7) | 52/54 (96.3) |
0, no expression; 1, low expression; 2, moderate/high expression.
In sinonasal specimens, most samples (46/64; 71.9%) showed low ACE2 expression, and TMPRSS2 protein was expressed in 39/59 samples (66.1%) as analyzed by IHC (Fig. 1b; Table 2); 27 of those (69.2%) also showed co-expression with ACE2.
ACE2 is an interferon-regulated gene [34]. We therefore investigated whether the expression of ACE2 and the other molecules could be associated with different types of inflammation. In chronic rhinosinusitis, high CD147 expression levels correlated with the presence of eosinophils, whereas we did not observe such correlations for ACE2, TMPRSS2, or furin (online suppl. material S5). This is in contrast to findings by Wang et al. [35], who reported lower ACE2 expression in eosinophilic compared to noneosinophilic chronic rhinosinusitis, although this was shown on the mRNA level.
Furin and CD147 Proteins Are Consistently Expressed in Conjunctival and Sinonasal Tissues
Besides ACE2, it has been postulated that CD147 could be an alternative entry receptor for SARS-CoV-2 [25]. Moreover, furin has been described as an activating cofactor in viral entry [24]. In contrast to ACE2, CD147 was positive in all patients, with high expression levels in 47/59 (79.7%) conjunctival and 52/54 (96.2%) sinonasal samples, respectively (Fig. 1a, b; Table 2). In addition, in the sinonasal tract, CD147 was significantly (p = 0.02) more expressed in tissues with higher eosinophil counts (online suppl. material S5). Moreover, furin was detectable at low to high levels in most patients, with only 6/54 (11.1%) of conjunctival and 5/52 (9.6%) of sinonasal specimens being completely negative, respectively (Table 2).
For all the 4 molecules investigated, we did not find any correlation in their expression in conjunctival or sinonasal samples with respect to patient age, sex, anatomic sinus, or prior history of malignancy (data not shown). In summary, these data indicate that ACE2 and TMPRSS2, the main entry molecules for SARS-CoV-2, are expressed at low protein levels in a subset of conjunctival and sinonasal tissues, with detectable co-expression in more than two-thirds of positive samples.
ACE2 and TMPRSS2 Proteins Are Only Focally Expressed in Epithelial Cells of the Lower Respiratory Tract
SARS-CoV-2 enters human cells via ACE2 receptors [21, 36], but the exact localization of virus entry is a matter of debate, and the precise pathophysiology of alveolar damage in COVID-19 is still under investigation [37]. In line with Aguiar et al. [18] and Hikmet et al. [19], in our cohort, ACE2 was mostly undetectable by IHC in bronchiolar and alveolar epithelium, and only 7/68 (10.3%) of bronchiolar and 1/75 (1.3%) of alveolar samples showed low-level expression, respectively (Fig. 1c, d; Table 3). Whereas TMPRSS2 was expressed at low levels in almost half of all bronchiolar samples (28/62; 45.2%), this molecule was absent in most alveolar epithelial cells and only detected in 9/70 (12.9%) samples (Fig. 1c, d; Table 3). Interestingly, and in contrast to observations by Ortiz et al. [17], in our cohort, ACE2 was strongly expressed in alveolar macrophages in all samples studied, which was also observed for TMPRSS2, furin, and CD147 (online suppl. material S4). Furthermore, ACE2 was more frequently expressed in bronchiolar epithelium in patients with an associated pulmonary malignancy (p = 0.012; data not shown).
Table 3.
Receptor protein expression in bronchioli and alveoli (%)
| Median | Negative, n/n (%) | Low, n/n (%) | High, n/n (%) | |
|---|---|---|---|---|
| Bronchioli | ||||
| ACE2 | 0 | 61/68 (89.7) | 7/68 (10.3) | 0/68 |
| TMPRSS2 | 0 | 34/62 (54.8) | 28/62 (45.2) | 0/62 |
| Furin | 1 | 27/64 (42.2) | 37/64 (57.8) | 0/64 |
| CD147 | 1 | 23/66 (34.8) | 23/66 (34.8) | 20/66 (30.3) |
|
| ||||
| Alveoli | ||||
| ACE2 | 0 | 74/75 (98.7) | 1/75 (1.3) | 0/75 |
| TMPRSS2 | 0 | 61/70 (87.1) | 9/70 (12.9) | 0/70 |
| Furin | 0 | 57/72 (79.2) | 15/72 (20.8) | 0/72 |
| CD147 | 0 | 51/72 (70.8) | 17/72 (23.6) | 4/72 (5.6) |
0, no expression; 1, low expression; 2, moderate/high expression.
In contrast to ACE2 and TMPRSS2, furin was detectable in the respiratory epithelium of many bronchioli (37/64, 57.8%), although only at low levels. In alveoli, low-level furin expression was only detected in a minority of samples (15/72, 20.8%) (online suppl. material S3d and Table 3). Interestingly, furin expression in bronchioli was higher in elderly patients: 25/36 (69.4%) of patients >60 years old showed detectable expression compared to patients ≤60 years old (12/28, 42.9%; p = 0.011). Moreover, the majority (16/23; 69.6%) of patients with concomitant pulmonary adenocarcinoma showed at least low furin expression levels in nontumorous bronchiolar epithelium (data not shown). Furthermore, furin was significantly more often detectable in bronchioli and alveoli of patients with prior history of malignancy (bronchioli: p = 0.005; alveoli: p = 0.066; data not shown). TMPRSS2 and CD147 showed a trend toward higher expression in alveoli of patients with prior history of malignancy (TMPRSS2: p = 0.074; CD147: p = 0.062).
We also investigated whether the expression of the 4 molecules in bronchioli correlated with alveolar expression. ACE2 (ρ = 0.433; p < 0.001), TMPRSS2 (ρ = 0.277; p = 0.032), furin (ρ = 0.463; p < 0.001), and CD147 (ρ = 0.377; p = 0.002) expression correlated significantly in bronchioli and alveoli (data not shown). In summary, our results indicate that ACE2, the main SARS-CoV-2 receptor protein, is only focally expressed in a minority of lower respiratory tract samples.
Discussion
Here, we systematically investigated protein expression of the most important SARS-CoV-2 receptor proteins in equally sized cohorts among possible viral entry sites in humans. In the light of previously described conflicting results on ACE2 expression in the human respiratory tract [15, 16, 17, 18, 19], we aimed at clarifying expression patterns. We found absent or only low ACE2 expression in the vast majority of cases in all investigated anatomic locations of the human airway tract. In accordance with the previous literature, cilia and alveolar macrophages expressed ACE2 most consistently [15, 38].
The ACE2 receptor has so far mainly been validated as the principal entry receptor on the mRNA level with high expression in respiratory cells of the lower airway tract [39]. The significance of these findings however is controversial [40]. Few studies assessed protein expression via IHC, with conflicting results. ACE2 is described to be expressed at low levels in the lower airway tract in general [15, 17, 41] but shows higher expression levels in other epithelial cell types, such as prostatic ductal epithelium (online suppl. material S1) [42]. Consistent with the previous literature, here, in the majority of cases, ACE2 was not expressed in lung parenchyma, i.e., type I pneumocytes [18, 19]. While Aguiar et al. [18] described diffuse staining of TMPRSS2 in airway epithelium of the lung, we only detected focal and low-level expression.
We confirm the previously described ACE2 expression in cilia [15, 16], here in the metaplastic conjunctival and respiratory nasal epithelium, and lack of expression in goblet cells. Here, 40.4% of conjunctival specimens showed low ACE2 expression, mainly in apical cilia of metaplastic ciliated epithelium. Primary cilia have been shown to play a role in murine surface ocular epithelium homeostasis, and patients with severe ciliopathies suffer from several ocular surface diseases [43]. However, squamous conjunctival epithelium was negative for ACE2, which is in line with findings by Lange et al. [44]. In the majority of sinonasal specimens, we found continuous low-level ACE2 expression in the apical respiratory epithelium, which confirms previous findings [15, 16] but is in contrast to other studies with significantly smaller cohorts that failed to detect ACE2 expression [19] or reported only focal apical expression [17].
In contrast to Ortiz et al. [17], we observed intense ACE2, TMPRSS2, furin, and CD147 expressions in alveolar macrophages. Macrophages are discussed as potential viral target cells [45]. M1 alveolar macrophages are capable of ingesting SARS-CoV-2 via phagocytosis, and ACE2 seems to play a role in this process [38, 46]. Furthermore, ACE2 expression in splenic macrophages and their vulnerability to SARS-CoV-2 infection have been demonstrated [47].
The role of children and adolescents in the spread of the pandemic has been a matter of debate [48, 49]. Here, minors (≤18 years, n = 15) only showed low ACE2 expression in sinonasal tissues (6/8, 75%). No expression was found in conjunctival tissue (0/4, 0%), bronchi, and alveoli (0/3, 0%, respectively). TMPRSS2 expression was, in general, negative or low as well. Importantly, detectable levels of expression of the 4 molecules were not correlated with patient sex, indicating that other factors are more important for the observed sex bias toward SARS-CoV-2 infection and severe COVID-19 [50].
SARS-CoV-2 spike protein exhibits cleavage sites for the enzyme furin, which has been postulated as a viral entry cofactor [4, 51, 52]. The absence of these cleavage sites reduces viral pathogenicity [53], and furin inhibition was suggested as a potential therapeutic strategy in COVID-19 [54]. In our cohorts, furin staining intensities were almost uniformly low and were mostly negative in lung alveoli. Moreover, we observed a trend toward higher expression levels in patients with a medical history of malignancy. High furin expression has been described in several cancer entities including lung carcinoma [55]; future studies are needed to investigate the role of furin in the pathogenesis of COVID-19 in cancer patients.
Finally, CD147 has been proposed as a viral entry receptor and a target for COVID-19 treatment [25, 56], but its expression has mainly been investigated at the mRNA level [57]. However, other authors postulated no relevance for CD147 as a viral entry facilitator [26, 27]. We observed heterogeneous CD147 expression in the tissues analyzed, which is consistent with previous results [18], with highest expression levels in conjunctival and sinonasal tissues. The exact role of CD147 in COVID-19 remains elusive and should be investigated further.
Limitations
This study has several limitations. First, the molecules investigated were only stained using a single antibody each. It was reported before that different antibodies against these molecules can have different staining patterns in the respiratory tract [15, 19]. Moreover, antibody lots may show inconsistencies over time, leading to staining discrepancies between studies. Second, in TMAs, only small areas of tissue are examined in contrast to full sections. This could lead to an underestimation of protein expression, especially if expression is only focal. We aimed at reducing this potential bias by including multiple TMA cores per specimen. Third, although in the same range as other studies, the size of our patient cohorts could be perceived as insufficient, especially when aiming at subgroup analyses. Finally, detailed patient history including smoking habits or previous infections was not available, precluding analyses linking those factors to expression levels of the proteins studied.
Conclusions
Our study confirms and extends previous reports. We found that ACE2 was mostly negative or only focally expressed at low levels in human tissues along possible viral entry routes. Where expressed, ACE2 mostly localized to the cilia and apical respiratory epithelial cells. Interestingly, ACE2 was almost uniformly negative in alveolar epithelia and only focally positive in bronchiolar respiratory epithelium in a minority of cases. This suggests that SARS-CoV-2 infection might occur primarily in the upper respiratory tract, implying important behavioral and clinical consequences. Finally, alveolar macrophages expressed all the investigated molecules, suggesting that this cell type could be directly infected by SARS-CoV-2 when inhaled as aerosols.
Statement of Ethics
The study was approved by the local Ethics Committee of the Canton of Bern, Switzerland (KEK 200/2014).
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
This work was supported by the Swiss Life Jubiläumsstiftung.
Author Contributions
K.B. and C.M.S. designed the study, analyzed the data, and wrote the manuscript. S.R. provided the scoring software and processed the data. J.A.G. performed IHC. M.W. and E.H. analyzed the data. All the authors revised the manuscript and approved its final version.
Data Availability Statement
Data are available upon reasonable request from the corresponding author.
Acknowledgments
The authors acknowledge the Translational Research Unit at the Institute of Pathology, University of Bern, Switzerland, for excellent technical support and Tissue Bank Bern (TBB) for providing the human tissue samples.
References
- 1.Bai Y, Yao L, Wei T, Tian F, Jin D-Y, Chen L, et al. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020 Apr 14;323((14)):1406–7. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johns Hopkins University Coronavirus COVID-19 global cases by the center for systems science and engineering (CSSE) [Internet] 2021. [cited 2021 May 25]. Available from: https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html?myid=3TGDV68B-3T9VW03K-SMHVE&utm_source=newsletter&utm_medium=email&utm_campaign=kk-2020-corona&utm_content=de#/bda7594740fd40299423467b48e9ecf6.
- 3.Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020 Aug 25;324((8)):782–93. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 4.Yuki K, Fujiogi M, Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin Immunol. 2020 Jun;215:108427. doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leisman DE, Ronner L, Pinotti R, Taylor MD, Sinha P, Calfee CS, et al. Cytokine elevation in severe and critical COVID-19: a rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir Med. 2020 Dec 1;8((12)):1233–44. doi: 10.1016/S2213-2600(20)30404-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van de Veerdonk FL, Netea MG, van Deuren M, van der Meer JW, de Mast Q, Brüggemann RJ, et al. Kallikrein-kinin blockade in patients with COVID-19 to prevent acute respiratory distress syndrome. Elife. 2020 Apr 27;9:e57555. doi: 10.7554/eLife.57555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fletcher-Sandersjöö A, Bellander B-M. Is COVID-19 associated thrombosis caused by overactivation of the complement cascade? A literature review. Thromb Res. 2020 Oct;194:36–41. doi: 10.1016/j.thromres.2020.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirchdoerfer RN, Cottrell CA, Wang N, Pallesen J, Yassine HM, Turner HL, et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016 Mar;531((7592)):118–21. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020 Mar;11((1)):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamming I, Timens W, Bulthuis MLC, Lely AT, Navis GJ, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203((2)):631–7. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia HP, Look DC, Shi L, Hickey M, Pewe L, Netland J, et al. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol. 2005 Dec;79((23)):14614. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scialo F, Daniele A, Amato F, Pastore L, Matera MG, Cazzola M, et al. ACE2: the major cell entry receptor for SARS-CoV-2. Lung. 2020 Dec 1;198((6)):867–77. doi: 10.1007/s00408-020-00408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samavati L, Uhal BD. ACE2, much more than just a receptor for SARS-COV-2. Front Cell Infect Microbiol. 2020;10:317. doi: 10.3389/fcimb.2020.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020 Apr 1;46((4)):586–90. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee IT, Nakayama T, Wu C-T, Goltsev Y, Jiang S, Gall PA, et al. ACE2 localizes to the respiratory cilia and is not increased by ACE inhibitors or ARBs. Nat Commun. 2020 Oct 28;11((1)):5453. doi: 10.1038/s41467-020-19145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakayama T, Lee IT, Jiang S, Matter MS, Yan CH, Overdevest JB, et al. Determinants of SARS-CoV-2 entry and replication in airway mucosal tissue and susceptibility in smokers. Cell Rep Med. 2021 Oct;2((10)):100421. doi: 10.1016/j.xcrm.2021.100421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ortiz ME, Thurman A, Pezzulo AA, Leidinger MR, Klesney-Tait JA, Karp PH, et al. Heterogeneous expression of the SARS-Coronavirus-2 receptor ACE2 in the human respiratory tract. EBioMedicine. 2020 Oct 1;60:102976. doi: 10.1016/j.ebiom.2020.102976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aguiar JA, Tremblay BJ-M, Mansfield MJ, Woody O, Lobb B, Banerjee A, et al. Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue. Eur Respir J. 2020 Sep 1;56((3)):2001123. doi: 10.1183/13993003.01123-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hikmet F, Méar L, Edvinsson Å, Micke P, Uhlén M, Lindskog C. The protein expression profile of ACE2 in human tissues. Mol Syst Biol. 2020 Jul 1;16((7)):e9610. doi: 10.15252/msb.20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glowacka I, Bertram S, Müller MA, Allen P, Soilleux E, Pfefferle S, et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85((9)):4122–34. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 Mar 5;181((2)):271–80.e8. doi: 10.1016/j.cell.2020.02.052. Available from: http://www.sciencedirect.com/science/article/pii/S0092867420302294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunst JD, Staerke NB, Pahus MH, Kristensen LH, Bodilsen J, Lohse N, et al. Efficacy of the TMPRSS2 inhibitor camostat mesilate in patients hospitalized with Covid-19-a double-blind randomized controlled trial. EClinicalMedicine. 2021;35:100849. doi: 10.1016/j.eclinm.2021.100849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann M, Hofmann-Winkler H, Smith JC, Krüger N, Arora P, Sørensen LK, et al. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine. 2021 Mar 1;65 doi: 10.1016/j.ebiom.2021.103255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bestle D, Heindl MR, Limburg H, Van Lam van T, Pilgram O, Moulton H, et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance. 2020 Sep 1;3((9)):e202000786. doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K, Chen W, Zhang Z, Deng Y, Lian J-Q, Du P, et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther. 2020 Dec 4;5((1)):283. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shilts J, Crozier TWM, Greenwood EJD, Lehner PJ, Wright GJ. No evidence for basigin/CD147 as a direct SARS-CoV-2 spike binding receptor. Sci Rep. 2021 Jan 11;11((1)):413. doi: 10.1038/s41598-020-80464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ragotte RJ, Pulido D, Donnellan FR, Gorini G, Davies H, Brun J, et al. Human basigin (CD147) does not directly interact with SARS-CoV-2 spike glycoprotein. bioRxiv. 2021 Jan 1; doi: 10.1128/mSphere.00647-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lytle NK, Ferguson LP, Rajbhandari N, Gilroy K, Fox RG, Deshpande A, et al. A multiscale map of the stem cell state in pancreatic adenocarcinoma. Cell. 2019 Apr 18;177((3)):572–86.e22. doi: 10.1016/j.cell.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.14(th) European congress on digital Pathology. J Pathol Inform. 2019 Nov 11;10:32. [Google Scholar]
- 30.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33((1)):159–74. [PubMed] [Google Scholar]
- 31.Desctools: tools for descriptive statistics [Internet] 2021. Available from: https://cran.r-project.org/package=DescTools.
- 32.Hui KPY, Cheung M-C, Perera RAPM, Ng K-C, Bui CHT, Ho JCW, et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir Med. 2020 Jul 1;8((7)):687–95. doi: 10.1016/S2213-2600(20)30193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng W, Bao L, Gao H, Xiang Z, Qu Y, Song Z, et al. Ocular conjunctival inoculation of SARS-CoV-2 can cause mild COVID-19 in rhesus macaques. Nat Commun. 2020 Sep 2;11((1)):4400. doi: 10.1038/s41467-020-18149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020 May;181((5)):1016–35.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang M, Bu X, Fang G, Luan G, Huang Y, Akdis CA, et al. Distinct expression of SARS-CoV-2 receptor ACE2 correlates with endotypes of chronic rhinosinusitis with nasal polyps. Allergy. 2021 Mar 1;76((3)):789–803. doi: 10.1111/all.14665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ni W, Yang X, Yang D, Bao J, Li R, Xiao Y, et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit Care. 2020 Jul 13;24((1)):422. doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osuchowski MF, Winkler MS, Skirecki T, Cajander S, Shankar-Hari M, Lachmann G, et al. The COVID-19 puzzle: deciphering pathophysiology and phenotypes of a new disease entity. Lancet Respir Med. 2021;9((6)):622–42. doi: 10.1016/S2213-2600(21)00218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lv J, Wang Z, Qu Y, Zhu H, Zhu Q, Tong W, et al. Distinct uptake, amplification, and release of SARS-CoV-2 by M1 and M2 alveolar macrophages. Cell Discov. 2021 Apr 13;7((1)):24. doi: 10.1038/s41421-021-00258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020 Apr 1;14((2)):185–92. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zamorano Cuervo N, Grandvaux N. ACE2: evidence of role as entry receptor for SARS-CoV-2 and implications in comorbidities. In: van de Veerdonk FL, van der Meer JW, editors. Elife. Vol. 9. 2020. Nov 9, p. e61390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matusiak M, Schürch CM. Expression of SARS-CoV-2 entry receptors in the respiratory tract of healthy individuals, smokers and asthmatics. Respir Res. 2020 Dec;21((1)):252. doi: 10.1186/s12931-020-01521-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sungnak W, Huang N, Bécavin C, Berg M, Queen R, Litvinukova M, et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020 May;26((5)):681–7. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grisanti L, Revenkova E, Gordon RE, Iomini C. Primary cilia maintain corneal epithelial homeostasis by regulation of the Notch signaling pathway. Development. 2016 Jun 15;143((12)):2160–71. doi: 10.1242/dev.132704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lange C, Wolf J, Auw-Haedrich C, Schlecht A, Boneva S, Lapp T, et al. Expression of the COVID-19 receptor ACE2 in the human conjunctiva. J Med Virol. 2020 Oct 1;92((10)):2081–6. doi: 10.1002/jmv.25981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gracia-Hernandez M, Sotomayor EM, Villagra A. Targeting macrophages as a therapeutic option in coronavirus disease 2019. Front Pharmacol. 2020;11:1659. doi: 10.3389/fphar.2020.577571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abassi Z, Knaney Y, Karram T, Heyman SN. The lung macrophage in SARS-CoV-2 infection: a friend or a foe? Front Immunol. 2020;11:1312. doi: 10.3389/fimmu.2020.01312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y, Feng Z, Diao B, Wang R, Wang G, Wang C, et al. The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes. medRxiv. 2020 Mar 31 [Google Scholar]
- 48.Lachassinne E, de Pontual L, Caseris M, Lorrot M, Guilluy C, Naud A, et al. SARS-CoV-2 transmission among children and staff in daycare centres during a nationwide lockdown in France: a cross-sectional, multicentre, seroprevalence study. Lancet Child Adolesc Health. 2021 Apr 1;5((4)):256–64. doi: 10.1016/S2352-4642(21)00024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viner RM, Mytton OT, Bonell C, Melendez-Torres GJ, Ward J, Hudson L, et al. Susceptibility to SARS-CoV-2 infection among children and adolescents compared with adults: a systematic review and meta-analysis. JAMA Pediatr. 2021 Feb 1;175((2)):143–56. doi: 10.1001/jamapediatrics.2020.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peckham H, de Gruijter NM, Raine C, Radziszewska A, Ciurtin C, Wedderburn LR, et al. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat Commun. 2020 Dec 9;11((1)):6317. doi: 10.1038/s41467-020-19741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020 Apr 1;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xia S, Lan Q, Su S, Wang X, Xu W, Liu Z, et al. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct Target Ther. 2020 Jun 12;5((1)):92. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson BA, Xie X, Kalveram B, Lokugamage KG, Muruato A, Zou J, et al. Furin cleavage site is key to SARS-CoV-2 pathogenesis. bioRxiv. 2020 Aug 26 [Google Scholar]
- 54.Wu C, Zheng M, Yang Y, Gu X, Yang K, Li M, et al. Furin: a potential therapeutic target for COVID-19. iScience. 2020 Oct 23;23((10)):101642. doi: 10.1016/j.isci.2020.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jaaks P, Bernasconi M. The proprotein convertase furin in tumour progression. Int J Cancer. 2017 Aug 15;141((4)):654–63. doi: 10.1002/ijc.30714. [DOI] [PubMed] [Google Scholar]
- 56.Ulrich H, Pillat MM. CD147 as a target for COVID-19 treatment: suggested effects of azithromycin and stem cell engagement. Stem Cell Rev Rep. 2020 Jun;16((3)):434–40. doi: 10.1007/s12015-020-09976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Radzikowska U, Ding M, Tan G, Zhakparov D, Peng Y, Wawrzyniak P, et al. Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy. 2020 Jun 4;75((11)):2829–45. doi: 10.1111/all.14429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available upon reasonable request from the corresponding author.