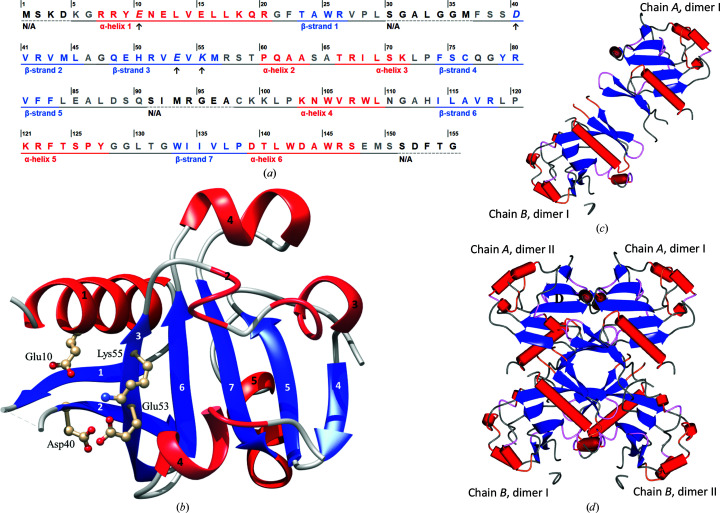
Figure 3.
(a) The secondary structure of the Hjc_15-6 monomer is displayed based on data from both chains A and B as described in UCSF Chimera and CCP4MG. (b) Hjc_15-6 protein monomer coloured by secondary structure with α-helices (red helices) and β-sheets (blue arrows) numbered in order from the N-terminus to the C-terminus. The image also includes the putative active site with the catalytic residues Glu10, Asp40, Glu53 and Lys55 labelled and presented in ball-and-stick representation (Fig. 3b was produced in UCSF Chimera with the native coordinates of PDB entry 7bnx chain B). (c) Hjc_15-6 protein displayed as a homodimer. Chains A and B are slightly differently modelled. (d) Hjc_15-6 protein displayed as a dimer of homodimers. In (c) and (d), Hjc_15-6 is also coloured by secondary structure, with α-helices as red tubes and β-sheets as blue arrows; both images were produced in CCP4MG.