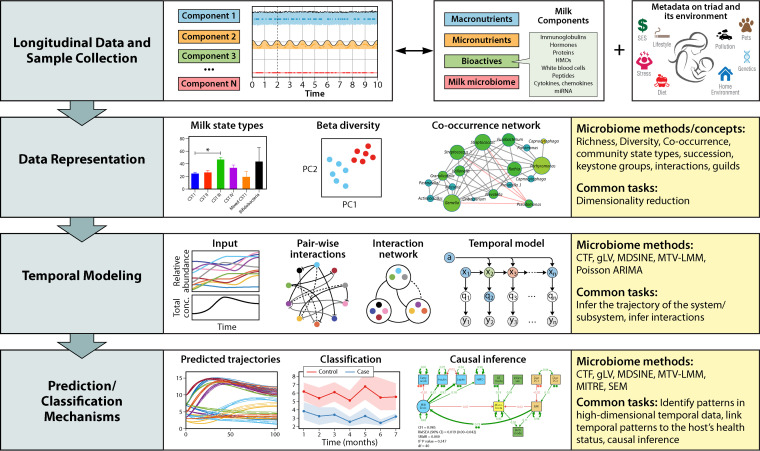
FIG 2.
Methodology for incorporating microbial community ecology and computational microbiome methods in the study of human milk. Longitudinal data collection involves collecting dense (frequent) human milk samples from women in tandem with high-dimensional metadata that capture the context (e.g., health, diet, environment) of the mother-milk-infant triad together with microbiome samples from both the mother and infant. Data representation entails applying computational methods or ecological concepts that summarize high-dimensional data, extracting important underlying structures in the data, and linking them to clinical outcomes. These concepts/methods include community state types, richness, diversity, co-occurrence networks, interactions, and more. Temporal modeling is inferring dynamical systems from milk time series data, using 4 steps as follows: (i) input is a time series of abundances of actors in the system or some lower-dimensional representation of the system over time (e.g., diversity over time); (ii) pairwise interaction network reflecting nonzero interaction coefficients in underlying dynamical systems model; (iii) interaction network with interaction module structure; and (iv) temporal model unrolled in time to explicitly show temporal dependencies. This schematic is inspired by methods developed for microbial dynamics (57). In predict/classify/elucidate mechanisms, by using data summaries extracted from the data representation methods/concepts as well as the temporal modeling, we can characterize the dynamics of human milk components, predict infant outcomes, and elucidate mechanisms underlying them. This can be done using statistical/probabilistic models, machine learning algorithms, mechanistic models that rely on ecology theory, and causal inference. Relevant microbiome methods include compositional tensor factorization (CTF) (63), generalized Lotka-Volterra (gLV) (57), Microbial Dynamical Systems INference Engine (MDSINE) (77), microbial temporal variability linear mixed model (MTV-LMM) (59), Poisson ARIMA (58), Microbiome Interpretable Temporal Rule Engine (MITRE) (64), and structural equation modeling (SEM) (78, 79).