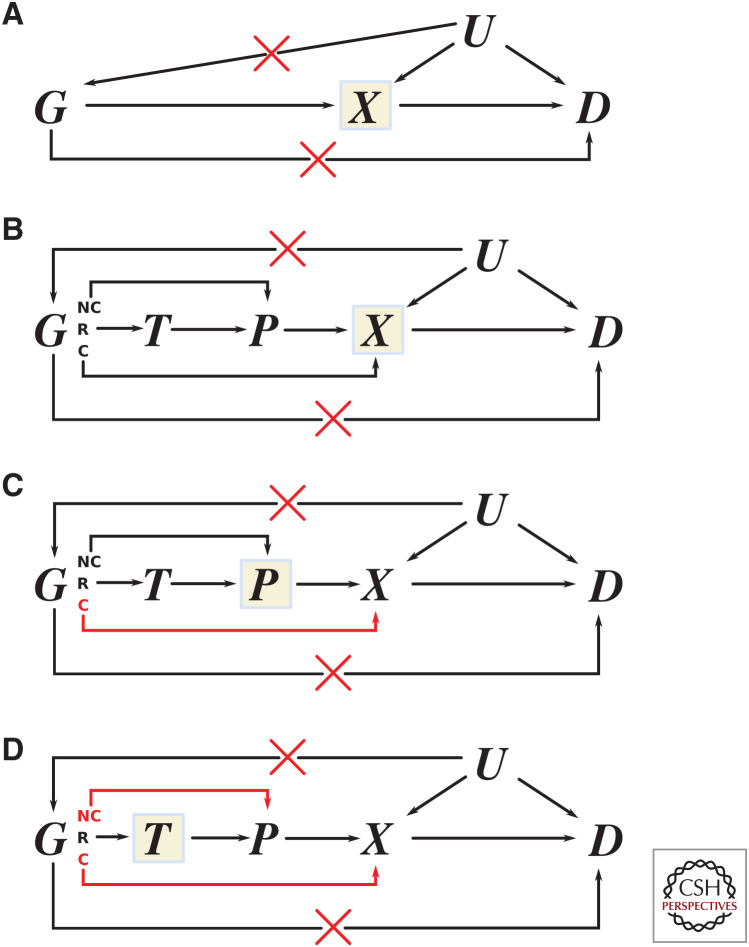
Figure 5.
Cis-acting variants with respect to different exposures and perceived heterogeneity. Multiple scenarios where cis-acting variants can impact the outcome but not via the exposure. (A) A conventional Mendelian randomization (MR) graph whereby instrumental variants in gene G are acting via biomarker exposure X on disease outcome D. (B) Mechanistically the same as part A but with greater resolution depicting potential (but unused) exposures of transcript level T and protein level P and with the instrumental genetic variants partitioned into noncoding (NC), regulatory (R), and coding (C) variants. Regulatory variants are more likely to impact the transcript level, whereas noncoding variants are more likely to impact protein level via translational efficiency and not the transcript level. Coding variants are more likely to impact on protein function/activity, which will alter downstream biomarker level but this effect will not be mediated via transcript or protein level. An exception to these assumptions is nonsense-mediated decay where aberrant insertion stop codons will lead to destruction of mRNA. (C) The exposure is changed from the biomarker to the protein encoded by gene G. (D) The exposure is changed from the level of protein to transcript level T. In C and D, pathways whereby instrumental genetic variants are impacting the outcome but not via the exposure are highlighted in red.