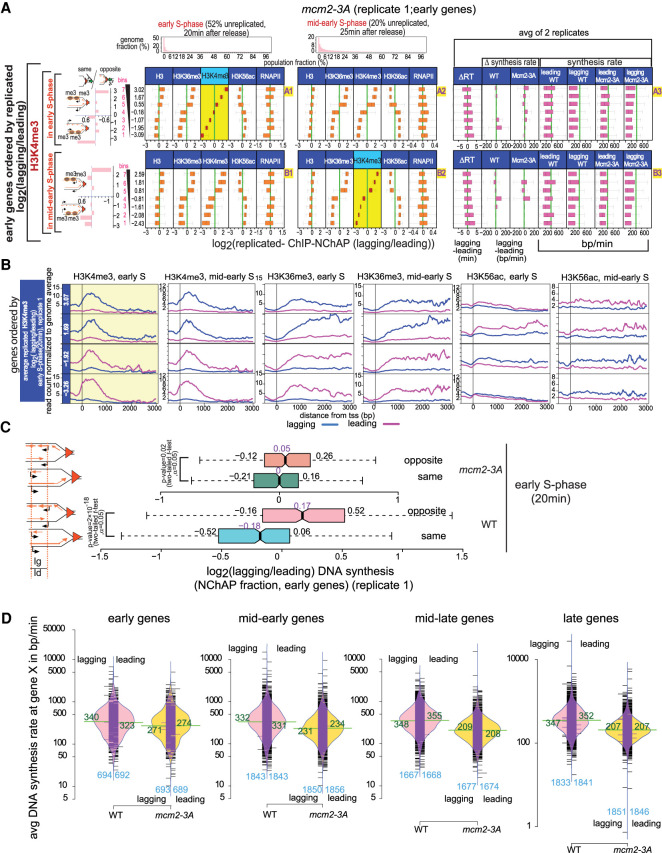
Figure 6.
Chromatin maturation in mcm2-3A cells. (A) Box plot distributions of lagging/leading ChIP-NChAP ratios for H3, H3K36me3, H3K4me3, H3K56ac, and RNAPII from early (column 1) and mid-early S phase (column 2) for early genes measured in the same culture of mcm2-3A mutant cells (biological replicate 1). The histograms on top show the distribution of genome read densities for each NChAP (replicated DNA) fraction at indicated time points in S phase. (Rows A and B) Genes have been sorted by decreasing lagging/leading occupancy of H3K4me3 in early S phase (row A) and mid-early S phase (row B), respectively (yellow background), and divided into seven bins (y-axis on the left). Box plot distribution of lagging/leading ratios (x-axis) for the chromatin features indicated in the header have been determined for each bin, as in Figure 5A. The bar graphs on the left show the “same” gene enrichment for gene bins indicated in the y-axis of each row on the right. Column 3 shows box plot distributions for, from left to right, the difference in replication timing (ΔRT) between the lagging and the leading strand for each gene in mcm2-3A cells; ΔDNA synthesis rates (lagging-leading) in WT and mcm2-3A cells for each gene in the bin; and average leading and lagging DNA synthesis rates in WT and mcm2-3A cells used to obtain the ΔDNA synthesis rates in Figure 5A for WT and in this figure for mutant cells. Synthesis rates for mcm2-3A were calculated as in Supplemental Figure S11 using replication timing from Supplemental Figure S10 and ROADs determined from NChAP fractions of the 20- and 25-min time points of two biological replicates (replicate 1 from Fig. 6A; replicate 2 from Supplemental Fig. S9A). (B) Average TSS-centered metagene profiles of ChIP-NChAP fractions indicated in the header from mcm2-3A cells (replicate 1) from gene bins from A sorted according to the average log2(lagging/leading) ratios for H3K4me3 in early S phase. Only the two bottom (bins 1,2) and top (bins 6,7) bins are shown. The value of the average ratio for each bin is indicated in the blue strip on the left. (C) Box plot distribution of DNA synthesis bias (log2[lagging/leading] of the NChAP fraction) for early replicating “opposite” and “same” genes in early S phase (20 min after release from G1 arrest) for WT replicate 1 (Fig. 5A) and mcm2-3A replicate 1 (Fig. 6A). (D) Bean plot of lagging and leading strand synthesis rate distribution (from Supplemental Figs. S10, S11) at early, mid-early, mid-late, and late genes for WT and mcm2-3A strains. The mean for each distribution is shown in green. The number of genes in each distribution is shown in blue on the bottom of the plot. The black bars represent individual data points. The bulk of synthesis rates in mcm2-3A are lower than in WT. Rates are constant in WT for all genes, but they gradually decrease as a function of replication timing in mcm2-3A mutants.