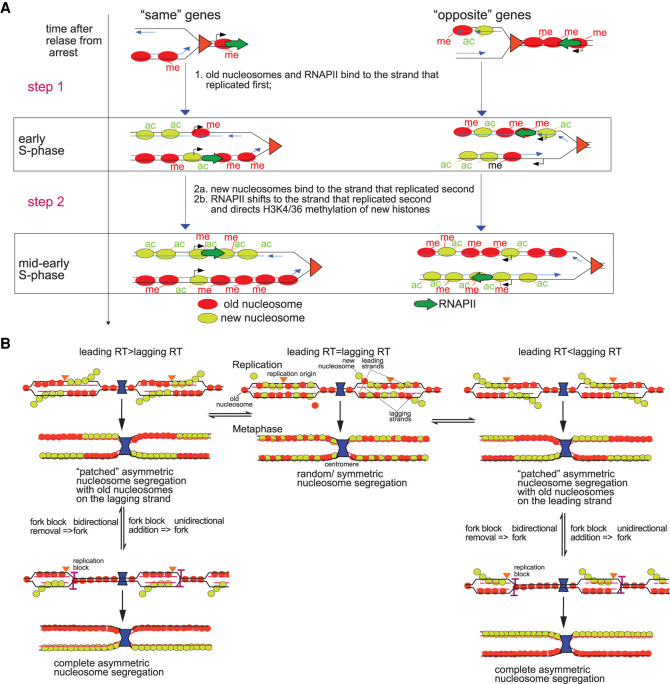
Figure 7.
Differences in replication timing between lagging and leading gene copies shape the distribution of old and new nucleosomes along sister chromatids. (A) Model for chromatin assembly on daughter chromatids. Nucleosome deposition follows a two-step process. (Step 1) “Old” nucleosomes (red) and RNAPII (green arrow) bind first to the leading strand behind the fork while the lagging strand is still replicating when transcription and replication travel in the same direction. When transcription and replication travel in opposite directions, old nucleosomes and RNAPII are deposited on the lagging strand that replicated first. New nucleosomes (light green) will be incorporated into the strand that replicated first mostly at promoters and ends of genes through replication independent turnover, although some will outcompete old nucleosomes for binding to other sites in the CDS. (Step 2) When replication of the other strand catches up, it will be mostly populated by new nucleosomes (2a) and RNAPII will then “switch” from the early replicating strand to the late one and direct H3K4 and H3K36 methylation of new histones by Set1p and Set2p, respectively, on the second gene copy (2b). (B) Modulation of the replication timing of replicated gene copies determines the pattern of old and new nucleosome segregation.