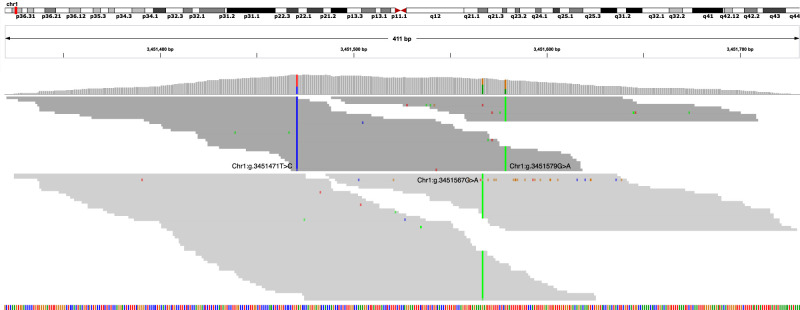
Figure 2.
Read pileup of HG003-HG004 tetraploid colored and grouped by supported haplotype. There are two distinct haplotypes (light and dark gray). The true genotypes for the three SNVs (Chr 1:g.3451471T > C, Chr 1:g.3451567G > A, Chr 1:g.3451579G > A) are AAAa, Aaaa, and AAAa. The variant allele read depths are 30/78 (38%), 38/64 (59%), and 20/58 (34%), respectively. GATK4 and FreeBayes both miscall the first two SNVs as AAaa, the most likely genotypes assuming binomially distributed allele observations. Octopus makes the correct calls because it phases all three SNVs, and the first haplotype (including the first and third SNVs) is supported by 74/114 (65%) of reads.