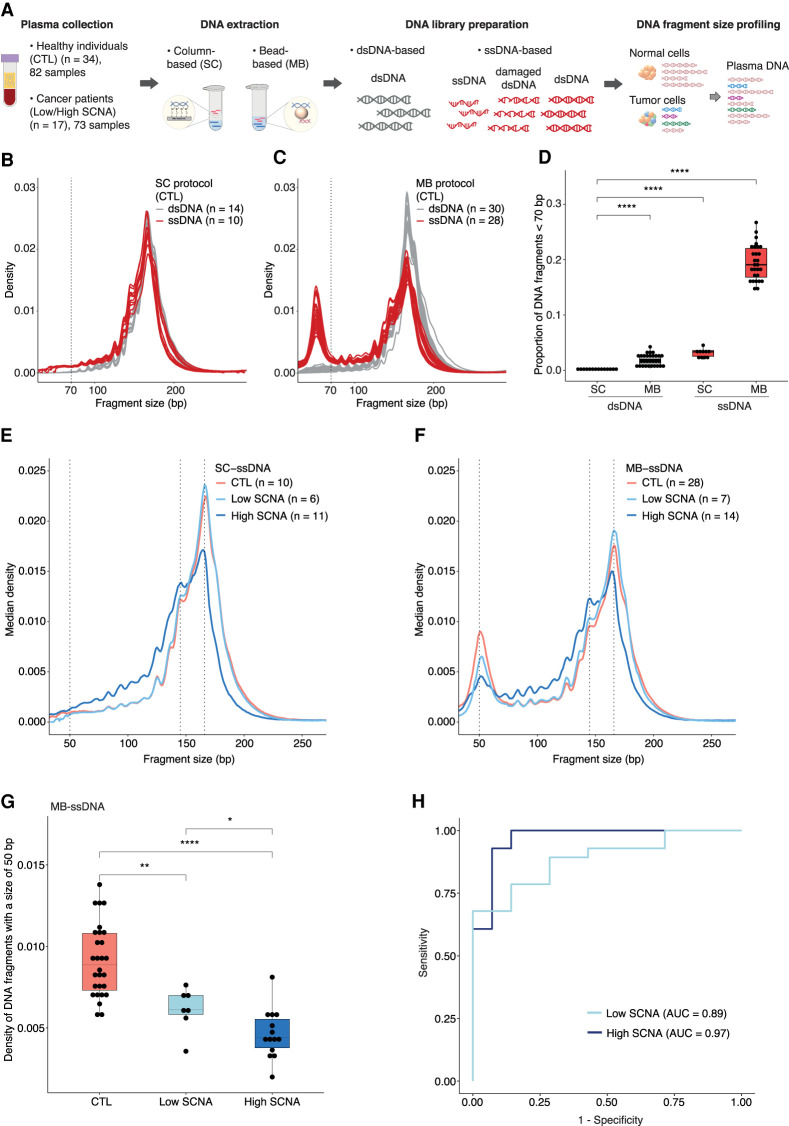
Figure 1.
Differences in the recovery of ultrashort cfDNA fragments using different DNA extraction and DNA library preparation protocols. (A) Plasma samples from healthy individuals and cancer patients were processed by column (SC)-based and magnetic bead (MB)-based DNA extraction methods, followed by dsDNA and ssDNA library protocols. The ssDNA library protocol recovers ssDNA and damaged dsDNA molecules, along with intact dsDNA, whereas the dsDNA library protocol recovers only intact dsDNA molecules. Paired-end sWGS data were generated from those libraries and used to determine DNA fragment size distributions. (B,C) cfDNA fragment size distributions of plasma ssDNA and dsDNA libraries from healthy individuals processed by SC (B) and MB (C) protocols. The vertical dashed line indicates a DNA fragment size of 70 bp. (D) Proportion of fragments shorter than 70 bp in plasma ssDNA and dsDNA libraries from healthy individuals. (E,F) Median fragment size distributions of cfDNA from plasma samples of patients with advanced cancers, divided into two groups: low SCNA (t-MAD < 0.019) and high SCNA (t-MAD ≥ 0.019), and analyzed by SC-ssDNA (E) and MB-ssDNA (F). Vertical dashed lines correspond to DNA fragment sizes of 50, 145, and 166 bp. (G) Density of fragments found at 50 bp in plasma samples from healthy individuals and cancer patients with low and high SCNA, data generated using the MB-ssDNA protocol. (H) ROC curve analysis of fragment density at 50 bp, generated using the MB-ssDNA protocol, for discriminating cancer patients from the healthy individuals. Wilcoxon rank-sum test: (*) P < 0.05; (**) P < 0.01; (****) P < 0.0001.