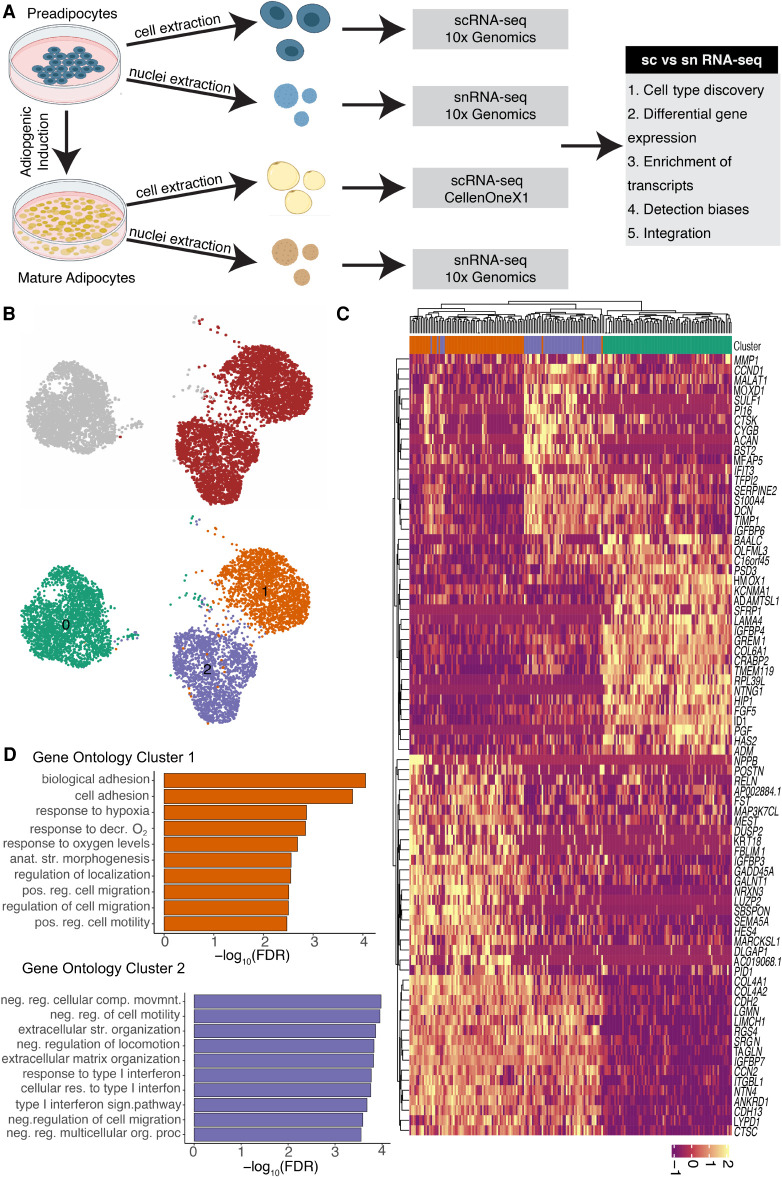
Figure 1.
scRNA-seq reveals transcriptional and compositional landscapes of white and brown preadipocytes. (A) A schematic representation of scRNA-seq versus snRNA-seq characterizations performed in our study. Comparisons were performed at both the preadipocyte stage and the mature adipocyte stage using an in vitro system of human preadipocytes. For the cell culture maintenance and differentiation protocol, see the Supplemental Methods. (B) UMAP visualization of white and brown preadipocytes either annotated manually to reflect the sample of origin (top panel) or annotated based on unsupervised clustering (bottom panel); 2578 white and 4055 brown cells were detected. Of these cells, 2558 were in cluster 0, 2062 in cluster 1, and 2013 in cluster 2. (C) Heat map of the top 20 differentially expressed genes between white and brown preadipocytes, as well as cluster 1 and cluster 2 preadipocytes based on log fold change values. Both genes (rows) and cells (column) underwent unsupervised clustering, with the top row reflecting cluster assignment as in B. (D) Top 10 Gene Ontology biological processes terms enriched in brown preadipocyte cluster 1 (top panel) and cluster 2 (bottom panel). (decr.) Decreased, (anat. str.) anatomical structure, (pos. reg.) positive regulation, (neg. reg) negative regulation, (str.) structural.