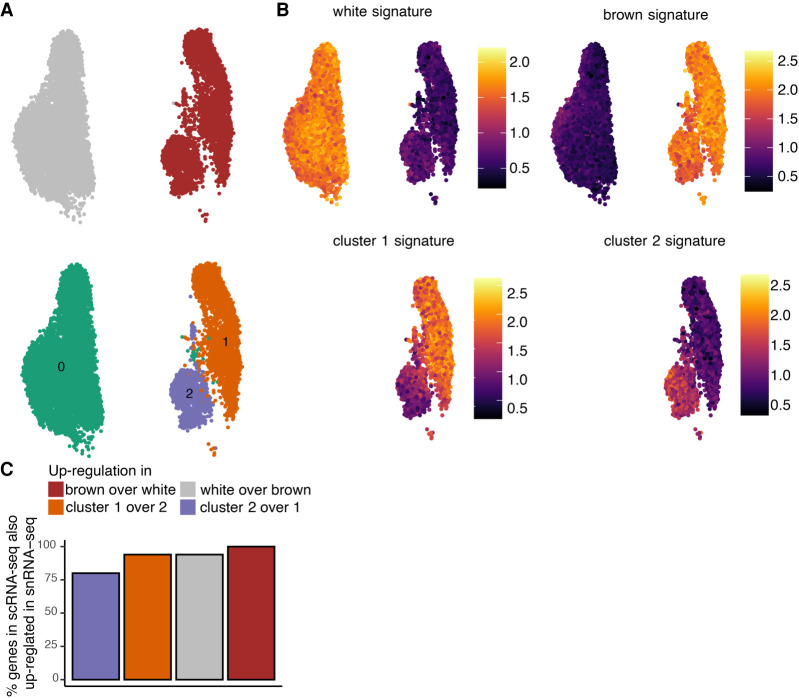
Figure 2.
snRNA-seq identifies the same preadipocyte populations as scRNA-seq and detects biologically relevant differential expression. (A) UMAP visualization of white and brown preadipocytes either annotated manually to reflect the sample of origin (top) or annotated based on unsupervised clustering (bottom); 6556 white and 3891 brown nuclei were detected. Of these nuclei, 6578 were in cluster 0, 2716 in cluster 1, and 1153 in cluster 2. (B) Heat map of transcriptional signature scores for white preadipocyte (top left), brown preadipocyte (top right), brown preadipocyte cluster 1 (bottom left), and brown preadipocyte cluster 2 (bottom right) as plotted on the UMAP visualization of snRNA-seq data. (C) Bar plot of percentage of top 50 genes differentially enriched (DE) in the scRNA-seq data set that are also DE in the snRNA-seq data set. The top 50 genes were evaluated based on log fold change values using the scRNA-seq data set.