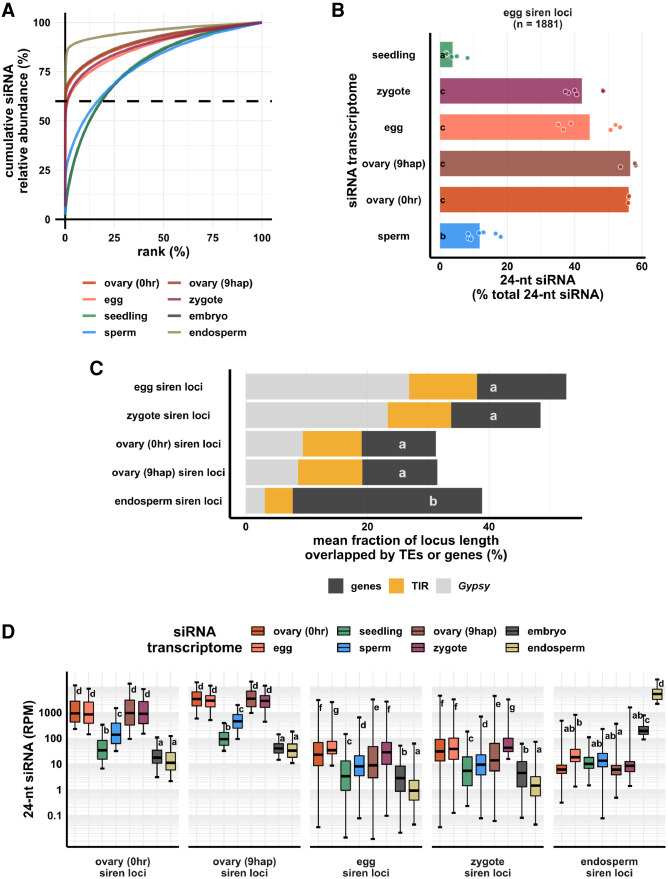
Figure 2.
Zygote siren loci are similar to siren loci detected in ovary and egg cell, and stably expressed between egg and zygote, but dissimilar to siren loci detected in endosperm. (A) x-axis is the rank order of siRNA loci. siRNA loci with highest siRNA abundances are ranked first. y-axis is cumulative relative abundance of siRNA in all siRNA loci. Axis values are scaled between 0% and 100%. 0.1% of siRNA loci accounted for 60% of siRNA reads in all siRNA loci in endosperm and ovary; 1% of siRNA loci accounted for 60% of siRNA reads in all siRNA loci in egg and zygote. (B) Bar plot showing relative abundances of 24-nt siRNAs at egg siren loci. Each data point is an siRNA transcriptome. Bar heights are averages. x-axis values are relative to total 24-nt siRNAs. (C) Stacked bar plots showing mean fraction of locus length overlapped by TEs or genes. (TIR) Terminal inverted repeat transposons, CACTA superfamily excluded; (Gypsy) Gypsy retrotransposons. (D) Box plots showing 24-nt siRNA relative abundances across siren classes across cell types. Middle lines are medians. Whiskers span 2.5th and 97.5th percentiles. Boxes span interquartile range. y-axis values are relative to per million total 24-nt siRNAs in each siRNA transcriptome. Letter grouping (α = 0.05) and P-values are based on Tukey tests. Embryo and endosperm data from Rodrigues et al. (2013). Seedling, gametes, and pre-fertilization ovary data from Li et al. (2020).