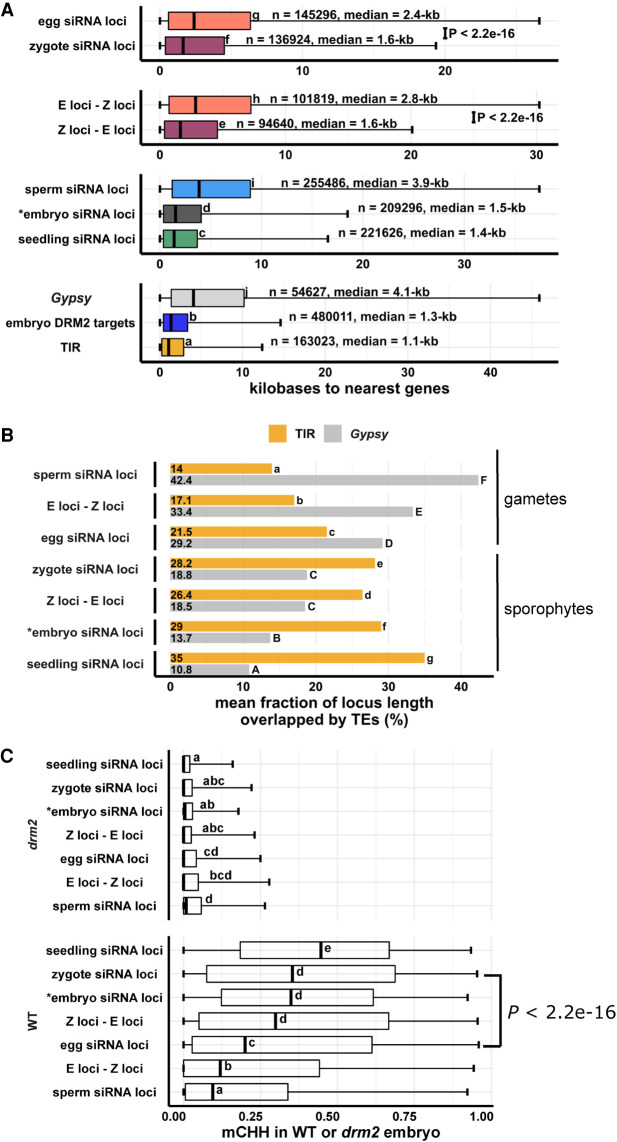
Figure 5.
Newly detected siRNA loci in zygote reset to the canonical siRNA profile and predict CHH methylation in embryo in an RdDM-dependent manner. (A) Box plots showing distance of siRNA loci to nearest genes. Middle lines are medians. Boxes span interquartile range. Whiskers span 2.5th and 97.5th percentiles. (B) Bar plots showing mean locus length overlapped by TIR transposon or Gypsy retrotransposons across siRNA loci categories. Statistical comparisons are made across siRNA loci categories within a TE length category. (C) Box plots showing CHH methylation level in mature wild-type and drm2 mutant embryos. Middle lines are medians. Boxes span interquartile range. Whiskers span 2.5th and 97.5th percentiles. E-Z loci: n = 101,841, Z-E loci: n = 94,591 (69% of all zygote siRNA loci). Letter groupings (α = 0.05) and P-values are based on Tukey tests. (*) Embryo siRNA data from Rodrigues et al. (2013), which was based on a single replicate. Seedling, gametes, and prefertilization ovary data from Li et al. (2020).