Abstract
Detection of Bordetella holmesii by a real-time PCR assay targeting IS481 of Bordetella pertussis is reported. Sequencing of IS481-specific PCR products from B. pertussis and B. holmesii isolates revealed sequence homology. Restriction fragment length polymorphism demonstrated a low copy number of IS481-like sequences in B. holmesii. These results, and culture of B. holmesii from patients with cough, suggest that the specificity and predictive value of IS481-based PCR assays for pertussis may be compromised.
Bordetella pertussis is the causative agent of whooping cough, an infectious disease that occurs worldwide with a high prevalence among young, unvaccinated infants (2, 3, 6, 19) and is recently resurgent in highly vaccinated populations. Related species, including B. parapertussis, B. bronchiseptica, and the more recently described B. holmesii (27), may also cause a pertussis-like syndrome in humans. Laboratory diagnosis of pertussis is traditionally based primarily on culture, which is highly specific but is maximally sensitive only in the initial phases of disease (10, 14). Furthermore, culturing depends on specimen quality and laboratory expertise and requires special media, extended incubation periods of 7 days or more, and confirmation by biochemical or antibody reagent tests. Diagnostic serology can be highly sensitive and more rapid than culture (14), but no serologic assay has been approved for diagnostic use in the United States because no diagnostic criterion has been widely accepted and no method has been validated between laboratories.
Thus, there is a need for more rapid and sensitive diagnostic methods that have high positive predictive value, especially in the early stages of disease. As is the case for other fastidious organisms, PCR offers an attractive alternative for detecting B. pertussis and B. parapertussis in clinical specimens (1, 4, 5, 7, 15, 17, 21, 26). Previously evaluated B. pertussis target regions include insertion sequences (IS), repeat elements, the pertussis toxin promoter region, the adenylate cyclase gene, and the porin gene. IS481 is present in the B. pertussis genome at 80 (22) to 100 (5, 18) copies, and PCR assays targeting IS481 have been evaluated for sensitivity and specificity in several laboratories over the last few years. Therefore, an internal region of IS481 was selected as the B. pertussis target (18) while a LightCycler (Roche Molecular Biochemicals, Mannheim, Germany) duplex PCR for B. pertussis and B. parapertussis was being developed. The previously described (5) forward primer BP-1 (5′ GAT TCA ATA GGT TGT ATG CAT GGT T 3′ and the slightly modified reverse primer BP-2 (5′ TTC AGG CAC ACA AAC TTG ATG GGC G 3′), corresponding to bp 12 to 36 and 192 to 167 (GenBank M22031) of IS481, respectively, were used for amplification. A pair of fluorescence-labeled hybridization probes, BP-HP-3 (5′ TCG CCA ACC CCC CAG TTC ACT CA-FAM 3′) and BP-HP-4 (5′ LC red 640-AGC CCG GCC GGA TGA ACA CCC-3′-phosphate), corresponding to bp 66 to 88 and 92 to 112 (GenBank M22031) of IS481, respectively, were used for the real-time detection of IS481-specific PCR products. Amplification mixtures contained 2 μl of 10× LightCycler FastStart DNA Master Hybridization Probes mix (Roche Molecular Biochemicals), 3 mM MgCl2, 0.5 μM concentrations of each primer oligonucleotide, 0.2 μM concentrations of each hybridization probe oligonucleotide, and 2 μl of template DNA in a final volume of 20 μl. Following an initial denaturation at 95°C for 10 min, the 50-cycle amplification profile consisted of heating at 20°C/s to 95°C with a 10-s hold, cooling at 20°C/s to 50°C with a 10-s hold, and heating at 20°C/s to 72°C with a 20-s hold.
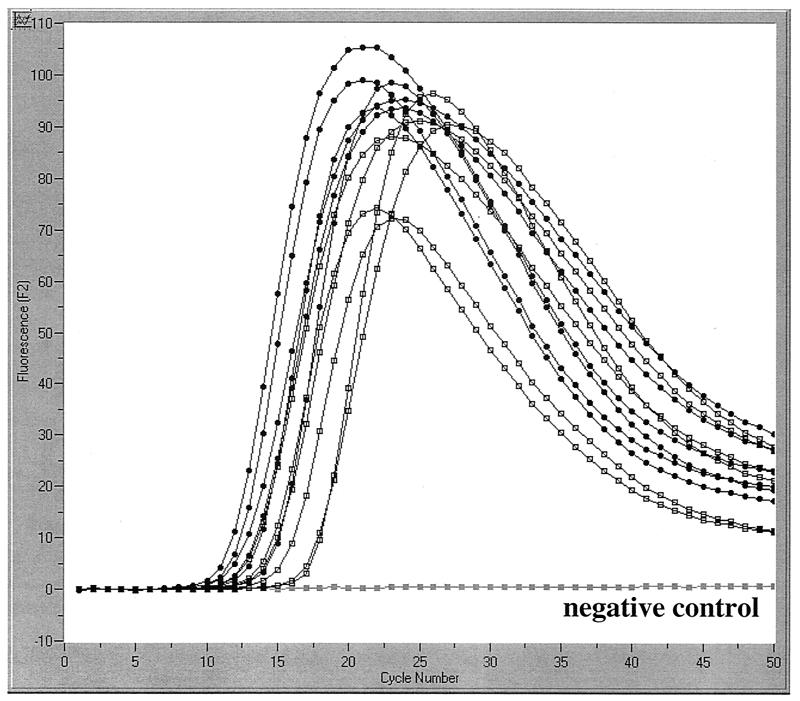
We evaluated the specificity of this primer-and-probe combination with the following: the type strains B. pertussis ATCC 11615 (American Type Culture Collection, Manassas, Va.) and B. holmesii ATCC 51541; isolates of B. pertussis (n = 15), B. parapertussis (n = 10), B. bronchiseptica (n = 2), B. holmesii (n = 5), and B. trematum (n = 1); and 80 gram-positive and -negative organisms other than Bordetella spp. (n = 80). A table of all isolates used will be provided upon request. Positive PCR results, as evidenced by probe fluorescence, were observed not only with the 16 B. pertussis isolates but also with B. holmesii type strain ATCC 51541 and all five B. holmesii clinical isolates (Fig. 1). No other isolates tested positive. The decrease in fluorescence intensity observed at cycle numbers above 25 is caused by the “hook effect,” when the number of amplicon molecules exceeds the number of fluorescence-labeled hybridization probe molecules present in the reaction mixture. This observation is in agreement with recent studies that detected B. holmesii genomic DNA by IS481-based PCR assays (8, 11, 28). We used direct sequencing of the respective 181-bp product amplified from the 1,053-bp IS481 coding sequence with all B. pertussis and B. holmesii isolates to demonstrate the molecular basis for this result: the two species share identical sequences within the amplified region of IS481. Sequence identity was further confirmed by sequencing cloned amplicons and 1,010-bp IS481 amplicons of B. pertussis and B. holmesii type strains (data not shown). Ambiguities were seen at nucleotide positions 133 (M) and 238 (Y) in all six B. holmesii isolates that may result from A/C or C/T variation, respectively, within the different IS481 alleles in the genome.
FIG. 1.
Evaluation of the IS481-specific LightCycler PCR assay with a set of six cultured strains of B. pertussis (type strain ATCC 11615 and five different patient isolates) and six cultured strains of B. holmesii (type strain ATCC 51541 and five different patient isolates [00, 91, 95, 96, and 98]; kindly provided by H. George, Massachusetts Dept. of Public Health). Amplicon curves represent B. pertussis (●) and B. holmesii (□) strains.
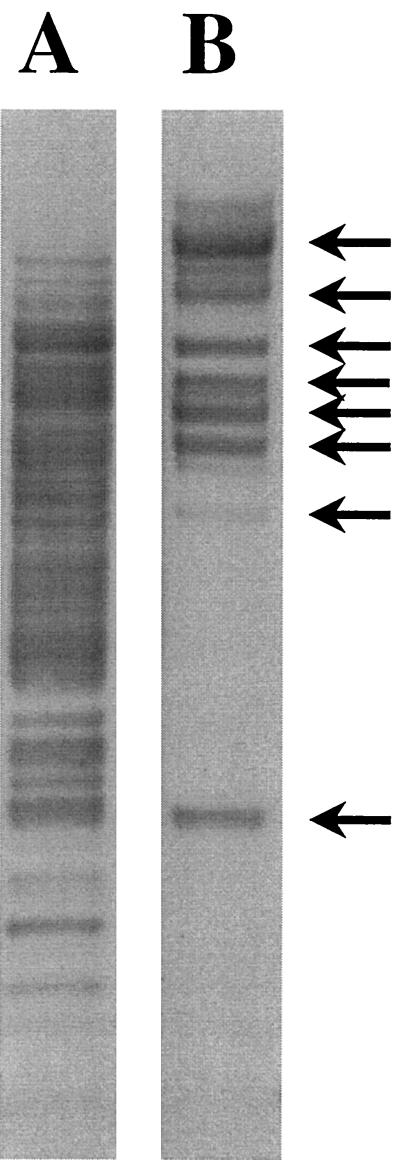
To further investigate the number and distribution of IS481-like sequences within the respective genomes of B. pertussis and B. holmesii, we used restriction fragment length polymorphism (RFLP) and Southern blot analysis. Bacterial strains were grown as described previously (24) and DNA extraction, digestion of chromosomal DNA with PvuII, and Southern blotting of restriction fragments were performed according to reported protocols (23, 25). The IS481 probe was labeled by direct incorporation of digoxigenin-dUTP during PCR (16) using primers BP-1 and BP-2. Southern blot membranes were exposed to X-ray films, and hybridizations were visualized by chemiluminescence. RFLP analysis demonstrated that IS481-specific sequences were present in high copy numbers (>50) in B. pertussis but in only 8 to 10 copies in B. holmesii (Fig. 2). This suggests that IS481 has been present in B. pertussis for a longer time than in B. holmesii. Although IS481 may have been inherited from an ancestor common to both B. pertussis and B. holmesii, recent evidence of a shared anatomic site (28) suggests the opportunity for the horizontal transfer of this element (22).
FIG. 2.
RFLP and Southern blot analysis for IS481 sequences. Chromosomal DNA of B. pertussis ATCC 11615 (A) and B. holmesii ATCC 51541 (B) was digested with PvuII, and the blot was hybridized with an IS481-specific probe. B. holmesii DNA fragments containing IS481-like sequences are indicated by arrows.
Amplification of targets within insertion sequences is attractive because their high genomic copy number (50 to 100 copies) allows increased assay sensitivity compared to single-copy targets. However, the insertion sequences of the eubacteria are transposable DNA elements capable of horizontal transfer across species and spontaneous elimination (29), characteristics that tend to decrease assay specificity and sensitivity, respectively. Due to the presence of IS481-like sequences in B. holmesii, a simple modification of PCR assay parameters or targeting of alternative IS481 primer sequences is unlikely to improve the specificity for B. pertussis.
The full clinical significance of false-positive B. pertussis IS481 PCR assay results due to detection of B. holmesii needs to be evaluated. Although B. holmesii is most often associated with septicemia in patients with underlying conditions (8, 9, 13, 20, 27), it recently accounted for <4% (32 of 868) of the Bordetella spp. cultured from nasopharyngeal secretions from Massachusetts patients with pertussis-like illness (12, 28). However, the clinical relevance and prevalence of B. holmesii in upper respiratory tract illness remain to be further investigated. Consequently, we recommend evaluation of other targets concurrent with the cautious use of IS481-based diagnostic PCR assays, including the interpretation and reporting of positive results.
Nucleotide sequence accession number.
The B. holmesii IS481-like insertion sequence was deposited in GenBank (accession no. AF349431).
Acknowledgments
We thank Katrin Kösters and Carl Heinz Wirsing von König for their active support and gratefully acknowledge the technical assistance of Stefan Lukas, Birgit Leppmeier, and Holger Melzl during the study.
REFERENCES
- 1.Backman A, Johansson B, Olsen P. Nested PCR optimized for detection of Bordetella pertussis in clinical nasopharyngeal samples. J Clin Microbiol. 1994;32:2544–2548. doi: 10.1128/jcm.32.10.2544-2548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Resurgence of pertussis: United States 1993. Morb Mortal Wkly Rep. 1993;42:952–960. [PubMed] [Google Scholar]
- 3.Deville J G, Cherry J D, Christenson P D, Pineda E, Leach C T, Kuhls T L, Viker S. Frequency of unrecognized Bordetella pertussis infection in adults. Clin Infect Dis. 1995;21:639–642. doi: 10.1093/clinids/21.3.639. [DOI] [PubMed] [Google Scholar]
- 4.Farell D J, Daggard G, Mukkur T K S. Nested duplex PCR to detect Bordetella pertussis and Bordetella parapertussis and its application in diagnosis of pertussis in nonmetropolitan Southeast Queensland, Australia. J Clin Microbiol. 1999;37:606–610. doi: 10.1128/jcm.37.3.606-610.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glare E M, Paton J C, Premier R, Lawrence A J, Nisbet L T. Analysis of a repetitive DNA sequence from Bordetella pertussis and its application to the diagnosis of pertussis using the polymerase chain reaction. J Clin Microbiol. 1990;28:1982–1987. doi: 10.1128/jcm.28.9.1982-1987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heininger U, Stehr K, Cherry J D. Serious pertussis overlooked in infants. Eur J Pediatr. 1992;151:342–343. doi: 10.1007/BF02113254. [DOI] [PubMed] [Google Scholar]
- 7.Heininger U, Schmidt-Schlapfer G, Cherry J D, Stehr K. Clinical validation of a polymerase chain reaction assay for the diagnosis of pertussis by comparison with serology, culture, and symptoms during a large pertussis vaccine efficacy trial. Pediatrics. 2000;105:E31. doi: 10.1542/peds.105.3.e31. [DOI] [PubMed] [Google Scholar]
- 8.Lind-Brandberg L, Welinder-Olsson C, Lagergard T, Taranger J, Trollfors B, Zackrisson G. Evaluation of PCR for diagnosis of Bordetella pertussis and Bordetella parapertussis infections. J Clin Microbiol. 1998;36:679–683. doi: 10.1128/jcm.36.3.679-683.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindquist S W, Weber D J, Mangum M E, Hollis D G, Jordan J. Bordetella holmesii sepsis in an asplenic adolescent. Pediatr Infect Dis J. 1995;14:813–815. [PubMed] [Google Scholar]
- 10.Loeffelholz M J, Thompson C J, Long K S, Gilchrist M J R. Comparison of PCR, culture, and direct fluorescent-antibody testing for detection of Bordetella pertussis. J Clin Microbiol. 1999;37:2872–2876. doi: 10.1128/jcm.37.9.2872-2876.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loeffelholz M J, Thompson C J, Long K S, Gilchrist M J R. Detection of Bordetella holmesii using Bordetella pertussis IS481 PCR assay. J Clin Microbiol. 2000;38:467. doi: 10.1128/jcm.38.1.467-467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazengia E, Silva E A, Peppe J A, Timperi R, George H. Recovery of Bordetella holmesii from patients with pertussis-like symptoms: use of pulsed-field gel electrophoresis to characterize circulating strains. J Clin Microbiol. 2000;38:2330–3333. doi: 10.1128/jcm.38.6.2330-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris J T, Myers M. Bacteremia due to Bordetella holmesii. Clin Infect Dis. 1998;27:912–913. doi: 10.1086/517173. [DOI] [PubMed] [Google Scholar]
- 14.Murphy T, Bisgard K, Sanden G N. Guidelines for the control of pertussis outbreaks, in press. Atlanta, Ga: Centers for Disease Control and Prevention; 2000. Diagnosis and laboratory methods. [Google Scholar]
- 15.Nygren M, Reizenstein E, Ronaghi M, Lundeberg J. Polymorphism in the pertussis toxin promoter region affecting the DNA-based diagnosis of Bordetella infection. J Clin Microbiol. 2000;38:55–60. doi: 10.1128/jcm.38.1.55-60.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reischl U, Rüger R, Kessler C. Nonradioactive labeling and high-sensitive detection of PCR products. Mol Biotechnol. 1994;1:229–240. doi: 10.1007/BF02921691. [DOI] [PubMed] [Google Scholar]
- 17.Schlapfer G, Cherry J D, Heininger U, Uberall M, Schmitt-Grohe S, Laussucq S, Just M, Stehr K. Polymerase chain reaction identification of Bordetella pertussis infection in vaccinees and family members in a pertussis vaccine efficacy trial in Germany. Pediatr Infect Dis J. 1995;14:209–214. doi: 10.1097/00006454-199503000-00008. [DOI] [PubMed] [Google Scholar]
- 18.Stibitz S. IS481 and IS1002 of Bordetella pertussis create a 6-base-pair duplication upon insertion at a consensus target site. J Bacteriol. 1998;180:4963–4966. doi: 10.1128/jb.180.18.4963-4966.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stojanov S, Liese J, Belohradsky B H. Hospitalization and complications in children under 2 years of age with Bordetella pertussis infection. Infection. 2000;28:106–110. doi: 10.1007/s150100050056. [DOI] [PubMed] [Google Scholar]
- 20.Tang Y W, Hopkins M K, Kolbert C P, Hartley P A, Severance P J, Persing D H. Bordetella holmesii-like organisms associated with septicemia, endocarditis, and respiratory failure. Clin Infect Dis. 1998;26:389–392. doi: 10.1086/516323. [DOI] [PubMed] [Google Scholar]
- 21.Van der Zee A, Agterberg C, Peeters M, Schellekens J, Mooi F R. Polymerase chain reaction assay for pertussis: simultaneous detection and discrimination of Bordetella pertussis and Bordetella parapertussis. J Clin Microbiol. 1993;31:2134–2140. doi: 10.1128/jcm.31.8.2134-2140.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van der Zee A, Groenendijk H, Peeters M, Mooi F R. The differentiation of Bordetella parapertussis and Bordetella pertussis from humans and animals as determined by DNA polymorphism mediated by two different insertion sequence elements suggests their phylogenetic relationship. Int J Syst Bacteriol. 1996;46:640–647. doi: 10.1099/00207713-46-3-640. [DOI] [PubMed] [Google Scholar]
- 23.Van Embden J D, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Loo I H, van der Heide H G, Nagelkerke N J, Verhoef J, Mooi F R. Temporal trends in the population structure of Bordetella pertussis during 1949–1996 in a highly vaccinated population. J Infect Dis. 1999;179:915–923. doi: 10.1086/314690. [DOI] [PubMed] [Google Scholar]
- 25.Van Soolingen D, Hermans P W, de Haas P E, Soll D R, van Embden J D. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wadowsky R M, Michaels R H, Libert T, Kingsley L A, Ehrlich G D. Multiplex PCR-based assay for detection of Bordetella pertussis in nasopharyngeal specimens. J Clin Microbiol. 1996;34:2645–2649. doi: 10.1128/jcm.34.11.2645-2649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weyant R S, Hollis D G, Weaver R E, Amin M F, Steigerwalt A G, O'Connor S P, Whitney A M, Daneshvar M I, Moss C W, Brenner D J. Bordetella holmesii sp. nov., a new gram-negative species associated with septicemia. J Clin Microbiol. 1995;33:1–7. doi: 10.1128/jcm.33.1.1-7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yih W K, Silva E A, Ida J, Harrington N, Lett S M, George H. Bordetella holmesii-like organisms isolated from Massachusetts patients with pertussis-like symptoms. Emerg Infect Dis. 1999;3:441–443. doi: 10.3201/eid0503.990317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuen L K, Ross B C, Jackson K M, Dwyer B. Characterization of Mycobacterium tuberculosis strains from Vietnamese patients by Southern blot hybridization. J Clin Microbiol. 1993;31:1615–1618. doi: 10.1128/jcm.31.6.1615-1618.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]