Figure 3 – T21 induced disruption of lamina associated domains (LADs) is associated with chromosomal introversion.
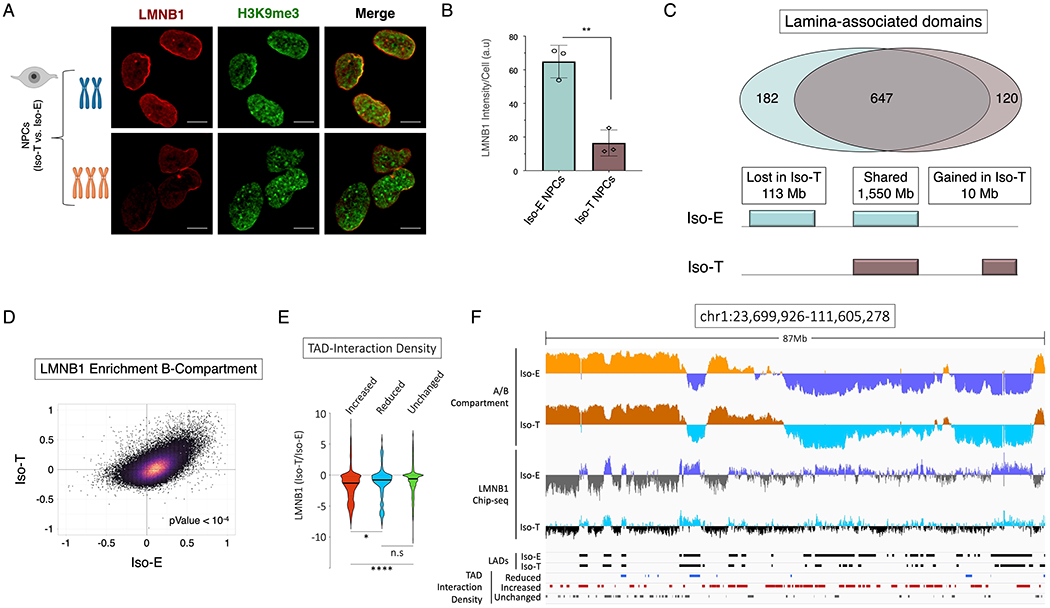
(A) Immunofluorescence of LMNB1 (red) and H3K9me3 (green) for the isogenic pair to assess LADs and heterochromatin distribution in NPCs. (B) Quantification of LMNB1 staining intensity in the isogenic pair of NPCs. Each dot on the histogram represents a replicate experiment of ~440 total nuclei analyzed for the isogenic pair of NPCs. (C) Venn diagram of LAD overlap between the euploid (Iso-E) and T21 (Iso-T) NPCs (intersect of replicates); bottom, schematic representation of genome coverage (base pairs) by LADs gained or lost as a consequence of T21 relative to euploid. (D) Scatter plot of LMNB1 enrichment over input in Iso-E (x-axis) and Iso-T (y-axis) of the genomic region in the B-compartment. (E) Violin plot of fold change of LMNB1 (Iso-T/Iso-E) of the differential (increased-ID (red), reduced-ID (blue) and unchanged-ID (green)) interaction density TADs. (F) IGV plot of A-compartment (Iso-E (yellow) and Iso-T (orange)) and B-compartment (Iso-E (blue) and Iso-T (light-blue)), LMNB1 ChIP-seq of Iso-E (blue) and Iso-T (light-blue), as well as the location of LADs (black) and the differentially interacting TADs (reduced-ID (blue), increased-ID (red) and unchanged-ID (gray)) an 87Mb region of chromosome 1.
