Figure 4 – T21 DEGs are distinctly separated into the A/B compartments in NPCs.
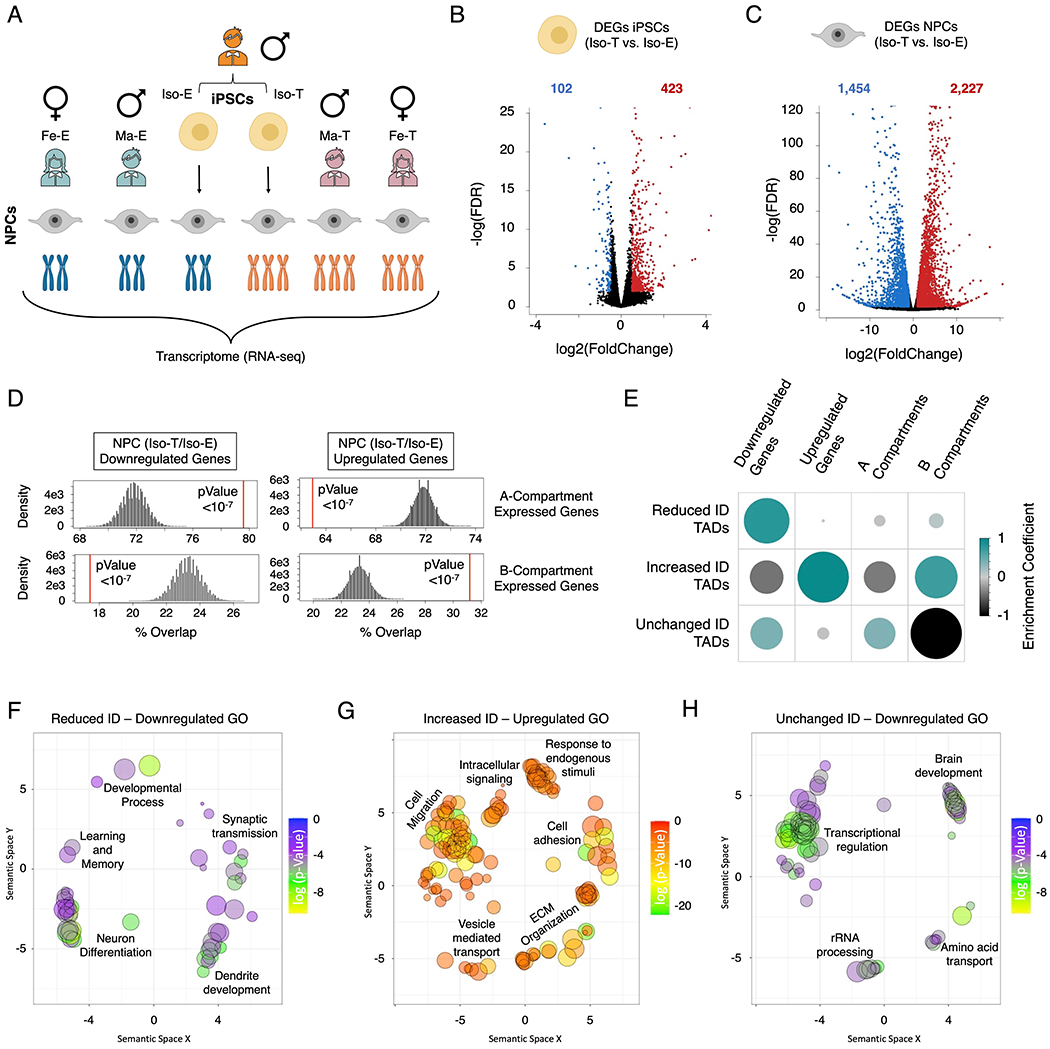
(A) Schematic of experimental design to assess the transcriptional consequences of T21 on the isogenic pair (Iso-E and Iso-T) of iPSCs and NPCs as well as NPCs derived from euploid (female (Fe-E) and male Ma-E)) and trisomic (female (Fe-T) and male (Ma-T)) individuals. (B, C) Volcano plots of the differentially expressed genes (DEGs) identified in the isogenic pair of iPSCs (B) and NPCs. (C). (D) Randomized permutation test between the downregulated and upregulated DEGs induced by T21 in NPCs with A/B compartments. Histogram represents the range of expected overlap percentage and the red line represents the observed percentage overlap. (E) Corrplot of the randomized permutation test between TADs with varying interaction densities (ID) with DEGs and A/B compartments. Color intensity (positive correlations are displayed in green and negative correlations in black) and the size of the circle are proportional to the enrichment coefficients. (F-H) Gene ontology visualized in semantic similarity-based scatterplots for downregulated genes associated with reduced-ID TADs (F), upregulated genes associated with increased-ID TADs (G), and downregulated genes localized within unchanged-ID TADs (H).
