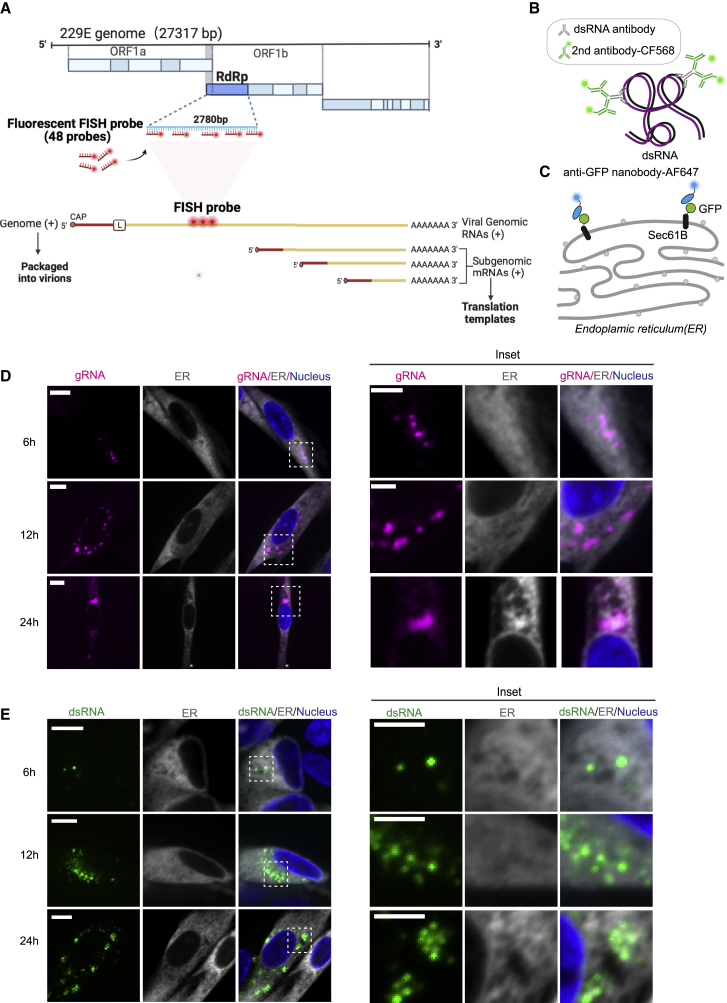
Figure 1.
Visualization of the positive-sense viral gRNA and dsRNA
(A) Top: 229E genomic construct map used for the detection of viral genome in fixed cells. Forty-eight FISH probes were designed to target gRNA by hybridizing with the RdRp-coding region, which is only present in the positive-sense gRNA and not in the sgRNAs. Bottom: Viral genome compared with sgRNAs, which do not contain complementary sequences to the FISH probes. The FISH probes are labeled by CF568 or AF647. Adapted from templates “Discontinuous Transcription” and “Remdesivir Active Molecule Interaction with SARS-CoV-2 RdRp”, by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender-templates (Hartenian et al., 2020).
(B) Scheme showing dsRNA targeted by anti-dsRNA antibodies in fixed cells. Each anti-dsRNA antibody recognizes 40 bp of dsRNA. A CF568-labeled secondary antibody is then introduced for visualization.
(C) Scheme showing the ER visualized in fixed cells via the expression of a single-pass ER membrane protein, GFP-Sec61B. An AF647-labeled anti-GFP nanobody is used to perform SR imaging of the ER.
(D) Representative confocal images of gRNA (magenta) in GFP-Sec61B-expressing fixed MRC5 cells (gray). Cells were infected with 0.2 multiplicity of infection 229E and fixed at 6-, 12-, and 24-h p.i. The nucleus is labeled with DAPI (blue). Scale bar, 10 μm. Right: insets of dotted boxes. Scale bar, 5 μm.
(E) Representative confocal images of dsRNA (green) in GFP-Sec61B-expressing fixed MRC5 cells (gray) at 6, 12, and 24 h p.i. with 0.2 multiplicity of infection 229E. The nucleus is labeled with DAPI (blue). Scale bar, 10 μm. Right: insets of dotted boxes. Scale bar, 5 μm