Figure 3.
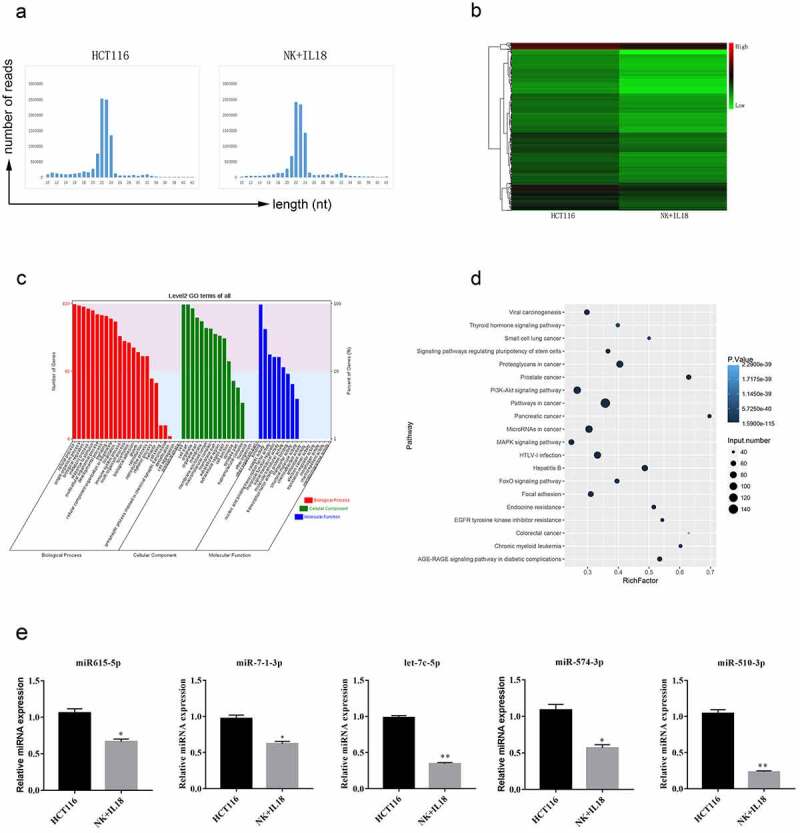
miRNA sequencing from HCT116 cells before and after NK cells coculturing and IL-18 treatment
a. Length of sequenced RNAs before and after NK cells coculturing and IL-18 treatment. miRNAs from both groups were distributed between 21 and 24 nt in length. b. Heatmap of differentially expressed miRNA. Twelve miRNAs had increased expression after treatment, whereas 697 miRNAs had decreased expression after treatment. (Difference in expression level is equal or greater than 1.5-fold was required to be considered as a change in expression.) c. Gene Ontology analysis of target mRNA of differentially expressed miRNAs. The majority of differentially expressed miRNAs are involved in biological processes. X-axis, biological pathways grouped as biological processes (red), cellular compartments (green), and molecular functions (blue); y-axis, count of miRNAs in each pathway. d. Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis. Target genes of the differentially expressed miRNAs are mainly involved in cancer, PI3K-AKT signaling, and function of microRNAs in cancer. e. Expression verification of hsa-miR615-5p, hsa-miR-7-1-3p, hsa-let-7c-5p, hsa-miR-574-3p, and hsa-miR-510-3p using qPCR. NK: Natural killer; IL-18: Interleukin 18; PI3K-AKT: Phosphatidylinositol 3-kinase and protein kinase B; qPCR: Quantitative polymerase chain reaction. All data with error bars are presented as mean ± SD, Student’s t test: * p < 0.05, ** p < 0.01.
