ABSTRACT
Lung cancer is a lethal cancer that threatens human health. Several studies have demonstrated the role of long non-coding RNAs (lncRNAs) in lung cancer. SOX21-AS1 is a newly discovered oncogenic lncRNA, but its molecular mechanism in lung cancer is not known. Here, the levels of SOX21-AS1, miR-24-3p, and PIM2 were examined in lung cancer and normal tissues. The relationships between miR-24-3p and SOX21-AS1 or PIM2 were predicted using bioinformatics tools and confirmed using a luciferase reporter assays. Colony formation, MTT, flow cytometry, and transwell assays were conducted to analyze cell proliferation, apoptosis, migration, and invasion abilities, respectively. Western blotting was used to measure PIM2 expression levels in cancer tissues and cells. SOX21-AS1 expression levels were high in lung cancer tissues and cells. In contrast, the amount of miR-24-3p bound to SOX21-AS1 was relatively low in cancerous tissues and cells. The knockdown of SOX21-AS1 decreased cell proliferation, activated apoptosis, and promoted cell migration and invasion. These effects were abolished by miR-24-3p inhibition. The oncogenic function of SOX21-AS1 mediated through targeting miR-24-3p was also demonstrated in animal models. PIM2 was targeted by miR-24-3p and showed increased levels in tumor tissues and cells. Furthermore, miR-24-3p overexpression inhibited the proliferation and promoted the apoptosis of lung cancer cells. In lung cancer cells, SOX21-AS1 negatively modulated the miR-24-3p/PIM2 axis to facilitate their proliferation, migration, and invasion. These findings offer a novel idea for future research on treating lung cancer at the molecular level.
KEYWORDS: Lung cancer, SOX21-AS1, miR-24-3p, PIM2
Graphical Abstract
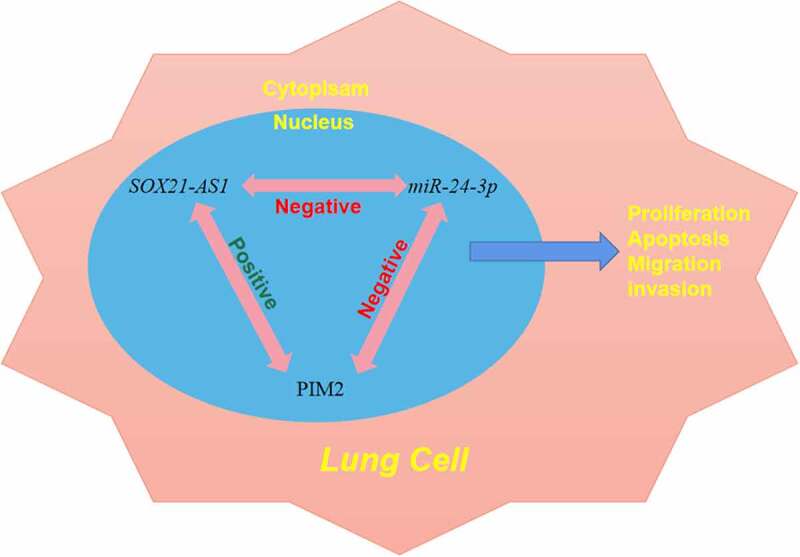
Introduction
Cancer is the leading cause of death worldwide and has become a serious threat to human health and social development [1]. Presently, deaths caused by lung cancer account for nearly 22.7% of deaths due to cancer, which is the biggest cause of death in China [2,3]. Lung cancer is generally classified as either small cell lung cancer (SCLC) or non-SCLC (NSCLC) based on histopathology, with 80––85% of cases being NSCLC [4,5]. Due to the invasiveness of lung cancer, some patients have local metastasis or infiltration at the initial consultation [6]. The 5-year survival rate of patients with tumor metastasis to other organs or systems is extremely low, but can be more than 40% for patients without tumor metastasis [7]. Studies have indicated that early diagnosis can improve the survival rate of lung cancer patients [8]. Therefore, exploring the molecular mechanism for the treatment of lung cancer has attracted much research interest.
Long non-coding RNAs (lncRNAs) are regarded as RNA fragments greater than 200 nt in length, without the ability to encode proteins [9,10]. Increasing evidence has shown that lncRNAs are ubiquitous in eukaryotes [11]. Recent studies have found that abnormal expression levels of lncRNAs are associated with tumor occurrence, metastasis, prognosis, and diagnosis. They can modulate the expression of protein-coding genes or regulate gene expression transcriptionally or post-transcriptionally, to govern different biological processes, such as proliferation, migration, invasion, apoptosis, and reprogramming of pluripotent stem cells [12–14]. The lncRNA, SOX21-AS1, is a newly discovered tumor-related lncRNA. It plays a vital role in a diverse range of cancers, including oral cancer, colorectal cancer, and hepatocellular carcinoma [15–17]. Only two studies have demonstrated an oncogenic role of SOX21-AS1 in lung cancer [18,19].
MicroRNAs (miRNAs) are RNA transcripts [20]. Increased levels of miR-24-3p have been shown to repress Bim to confer tamoxifen resistance in breast cancer cells [21]. TRIM11, which is directly targeted by miR-24-3p, accelerates the proliferation and apoptosis of colon cancer cells [22]. Moreover, miR-24-3p can interfere with lung adenocarcinoma progression through FGFR3 signaling [23]. PIM2, a downstream target of miR-24-3p, has already been shown to exert tumor-promoting effects in different carcinomas. Moreover, a decrease in PIM2 levels inhibits cell proliferation in liver cancer by modulating the cell cycle [24]. PIM2 and STAT3 form a positive feedback loop that regulates epithelial-to-mesenchymal transition in breast cancer [25]. However, the association between these genes has not been explored in the context of lung cancer.
Therefore, the aim of this study was to better understand the molecular mechanism underlying the role of SOX21-AS1 in lung cancer and to provide new ideas for research on lung cancer treatment. We hypothesized that SOX21-AS1 regulates the growth and metastasis of lung cancer cells via miR-24-3p/PIM2 axis.
Materials and methods
Clinical tissues
This study was approved by the Ethics Committee of The First Affiliated Hospital of Soochow University, and informed consent was obtained from all patients. Thirty matched cancerous and noncancerous lung tissue samples were obtained from patients at The First Affiliated Hospital of Soochow University. After surgery, the samples were frozen in liquid nitrogen.
Bioinformatics prediction
DIANA-LncBase v2 was used to predict the target miRNA of SOX21-AS1. TargetScan and miRDB were used to screen for common targets of miR-24-3p, and the results were further investigated using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses.
Cell culture
The lung cancer cell lines, H125, A549, NCI-H23, HCC827, and NCI-H1299, and normal human lung epithelial BEAS-2B cells were procured from ATCC (Manassas, VA, USA). Cells were grown in RPMI-1640 with 10% fetal bovine serum and 1% double antibody in an incubator at 37°C with 5% CO2. When the incubated cells reached 90% confluence, they were digested with trypsin and passaged.
Reverse transcription quantitative PCR
Total RNA was extracted from tissues and cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The purity and concentration of the RNA samples were determined using a NanoDrop ND-1000 spectrophotometer (Agilent, Santa Clara, CA, USA). The 260 nm/280 nm absorbance ratio of the RNA samples ranged from 1.8 to 2.1. RNA integrity was evaluated by 2% denatured agarose gel electrophoresis. Reverse transcription (RT) was performed using an RT-PCR kit (Promega, Madison, WI, USA). Quantitative (q) PCR was performed using an ABI PRISM 7500 Sequence Detection System (Life Technologies; Thermo Fisher Scientific, Waltham, MA, USA) and a SYBR Green Kit (Takara Biotechnology Co., Kusatsu, Japan). The levels of SOX21-AS1, miR-24-3p, and PIM2 were calculated using the ∆∆Cq method according to a previous study [26] .
Cell transfection
Vectors containing an SOX21-AS1 siRNA, miR-24-3p inhibitors or mimics, or PIM2, were transfected using Lipofectamine 2000 (Invitrogen), along with the corresponding negative controls (NCs), including an siRNA, miRNA inhibitor, and miRNA mimic NC, miR-NC inhibitor, miR-NC mimics, and pcDNA3.1. Culture medium without any added vector was used as a blank control. Six hours after transfection, the medium was replaced with fresh medium. Gene expression was assessed using RT-qPCR or western blotting to confirm the success of the transfection.
Colony formation assay
The Colony formation assay was performed as described by Lei et al. [27]. After cell transfection, 2 mL of 0.6% agarose was added to each well of a 6-well plate. Following the solidification of the agarose, the cells were suspended in 0.3% agarose was used to suspend the cells at 37°C, and then 2 mL of the cell suspension was quickly placed on the solidified agarose in the plate (2,000 cells/well). After incubation at 4°C for 10 min, the cells were cultured in a 37°C incubator for 14 d. The number of clones formed was then counted using a microscope (≥ 50 cells were regarded as one clone). Clone formation rate was calculated suing the following formula: (number of clones/number of inoculated cells) × 100.
MTT assay
MTT assay was conducted according to perious study [28]. Cells (1 × 105) were seeded in 96-well dishes to determine cell viability. At 0, 1, 2, and 3 d after transfection, 20 μL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, 5 μg/μL) was added. Dimethyl sulfoxide (150 μL/well) was then added after another 4 h of incubation and the absorbance of each group was measured at 570 nm for each group. The proliferation rates were calculated as the ratios of the absorbance values of the treatment group to the absorbance value of the control group were calculated as the proliferation rate.
Flow cytometric analysis
Two days after transfection, the cultured cancer cells were digested with trypsin and rinsed with phosphate-buffered saline. The concentration of the resuspended cells was adjusted to 1 × 104 cells/mL. The cells were then incubated with annexin V-FITC and propidium iodide in the dark for 15 min. The rate of apoptosis rate in each group was determined using flow cytometry (FACScan; BD Biosciences, Shanghai, China).
Transwell migration assay
The transwell migration assay was performed as reported by Omar et al [29]. Transfected cells were cultured for 48 h in Dulbecco’s modified Eagle medium (DMEM) supplemented with 0.1% bovine serum albumin (BSA) at a cell density of 1 × 106 cells/mL. Cells (100 μL) were inoculated into the upper transwell chamber (BD Biosciences), and 500 μL of DMEM containing 15% calf serum was added to the lower chamber. After 1 d of culture at 37°C with 5% CO2 at 37°C, cells on the upper layer were removed using a cotton swab. The filter membrane was then fixed with methanol for 5 min and stained with Giemsa stain for 15 min. Cells passing through the membrane were counted in five different fields under a light microscope (Zeiss Axio Observer; Zeiss, Oberkochen, Germany) at a magnification of 100× and the average value was calculated. Migration ability was calculated using the following formula: (1 – the average number of migrated cells in the experimental group/the number of migrated cells vs. those in the control group) × 100%.
Transwell invasion assay
The transwell invasion assay was performed as reported by Marshall et al. [30]. A Matrigel basement membrane matrix was used to investigate cell invasion. The density of the cells transfected for 48 h was adjusted to 3 × 105 cells/mL and 0.1 mL of the resulting cell suspension was added to each well of the upper transwell chamber containing serum-free medium. One milliliter of complete medium was then added to the lower insert. After 24 h, cells in the lower layer were fixed and stained with crystal violet.
Luciferase reporter assay
The luciferase reporter assay was conducted according to a previous study [31]. Wild-type (WT) and mutant (MT) fragments of SOX21-AS1 and PIM2 were separately amplified by RT-PCR and subcloned into pmirGLO expression vectors (Sangon Biotech, Shanghai, China). The above vectors were then co-transfected into 293 T cells with miR-24-3p mimics, miRNA mimics, and mimic NCs using Lipofectamine 2000. A dual-luciferase reporter assay system (TransGene, Beijing, China) was used to detect luciferase activity.
Tumor xenografts
Tumor xenografts was established as decribed by Lee et al [32]. After successful transfection, a cell suspension of 5 × 107 cells/mL was created via digestion with trypsin, centrifugation, collection, and counting. The cell suspension (0.2 mL) was injected subcutaneously injected into the middle dorsal armpit of nude mice. One week later, tumors appeared in situ. Tumor weight and size in each group (20 nude mice) were examined every fourth day. The longest diameter (a) and shortest diameter (b) were measured to calculate tumor volume (V), using the formula, V = πab2/6. After 30 d of observation, the nude mice were sacrificed and tumor tissues were immediately collected and weighed. The tumor inhibition rate was then calculated according to the following formula: tumor inhibition rate (%) = (1 – average tumor weight of treatment group/average tumor weight of control group) × 100%. Relative tumor volume (RTV) was calculated according to the formula: RTV = Vt/V0, where Vt is the final tumor volume, and V0 is the initial tumor volume. Relative tumor proliferation rate (%) was calculated as follows: (the RTV of the treatment group/the RTV of the control group) × 100%.
Western blotting
The western blotting was conducted according to a previous study [33]. Tissues or cells were treated with RIPA lysis buffer containing protease inhibitors. After centrifugation, the supernatant was heated in a water bath to denature the proteins. Following quantification using the bicinchoninic acid method, proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to a polyvinylidene difluoride membrane (Sigma, St Louis, MO, USA). The membranes were probed with primary antibodies against PIM2 (1:1,000; ab129193; Abcam, Cambridge, UK) and GAPDH (1:2,500; ab9485; Abcam) at 4°C for 12 h. Thereafter, the membranes were incubated with a secondary antibody against rabbit IgG (1:1,000; ab190475; Abcam) at room temperature for 1 h. Protein bands were detected using an enhanced chemiluminescence system (Super Signal West Pico Substrate, Thermo Fisher Scientific), followed by quantification using Image J software (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
Data from triplicate experiments were analyzed using SPSS software (version 22.0; IBM Corporation, Armonk, NY, USA) and expressed as means ± standard errors of the mean. Differences between two groups or among groups were evaluated using a Student’s t-test or analysis of variance. Statistical significance was set at P < 0.05.
Results
This study aimed to explore the role of SOX21-AS1 in the occurrence and development of lung cancer cells. We conducted the MTT, colony formation, flow cytometry, and transwell assays to detect the cell proliferation, apoptosis, migration and invasion in vitro, and explored the role of SOX21-AS1/miR-24-3p axis in the tumor xenografts in vivo.
SOX21-AS1 was expressed at high levels in lung cancer
The levels of lncRNA SOX21-AS1 in lung cancer and adjacent tissues were determined using RT-qPCR. The data demonstrated that SOX21-AS1 expression was markedly higher in cancer tissues than in non-cancer tissues (Figure 1(a)). Moreover, SOX21-AS1 levels were also examined in cancerous and normal cells. RT-qPCR analysis revealed that SOX21-AS1 levels were higher in A549, NCI-H23, H125, and HCC827 cells than in BEAS-2B cells (Figure 1(b)). A549 and HCC827 cells showed the highest levels of SOX21-AS1 among all the cancer cell lines tested. SOX21-AS1 expression levels were also significantly higher in patients with lung cancer.
Figure 1.
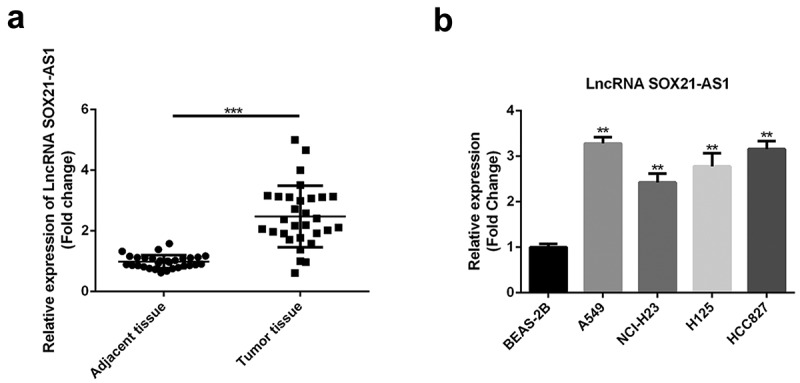
High expression levels of SOX21-AS1 in lung cancer
(a) SOX21-AS1 levels in lung cancer and adjacent tissues were determined by RT-qPCR. ***P < 0.001 vs adjacent tissue. (b) The expression of SOX21-AS1 in normal lung epithelial cells (BEAS-2B) and lung cancer cells (H125, A549, NCI-H23, HCC827, and NCI-H1299) was also analyzed by RT-qPCR. **P < 0.01 vs BEAS-2B cells.
SOX21-AS1 targeted miR-24-3p in lung cancer
Bioinformatics tools revealed that miR-24-3p was a potential target of SOX21-AS1. The binding area is shown in Figure 2(a). Therefore, we analyzed miR-24-3p levels in normal and cancerous tissues. Reduced levels of miR-24-3p were observed in cancerous tissues (Figure 2(b)). In addition, miR-24-3p levels were markedly decreased in cancerous cells compared to normal BEAS-2B cells (Figure 2(c)). To confirm the binding of SOX21-AS1 with miR-24-3p, a luciferase reporter assay was performed. The luciferase activity of the wild-type SOX21-AS1 construct was clearly decreased by overexpression of miR-24-3p (Figure 2(d)). In summary, we confirmed that miR-24-3p bound to SOX21-AS1.
Figure 2.
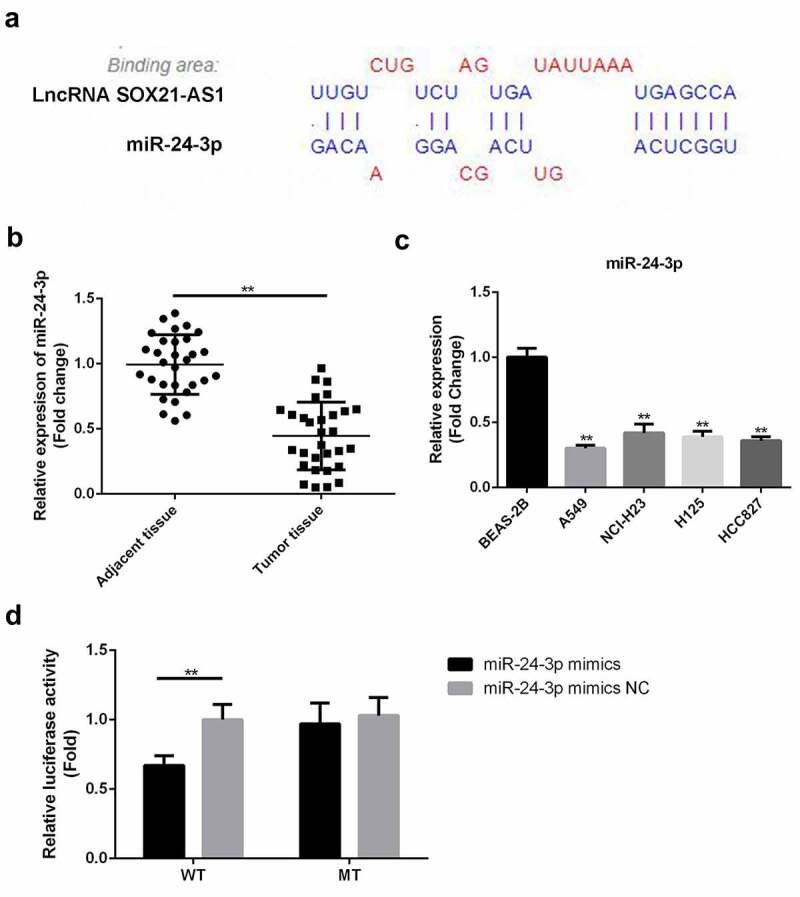
SOX21-AS1 targeted miR-24-3p in lung cancer
(a) The binding site between SOX21-AS1 and miR-24-3p as predicted by DIANA. (b) miR-24-3p levels in lung cancer and adjacent tissues were determined by RT-qPCR. **P < 0.01 vs adjacent tissue. (c) miR-24-3p expression levels in normal BEAS-2B cells and lung cancer cells were determined by RT-qPCR. **P < 0.01 vs BEAS-2B cells. (d) Luciferase reporter assays were used to assess the binding affinity between SOX21-AS1 and miR-24-3p. **P < 0.01 vs miR-24-3p mimic NC.
miR-24-3p inhibition reversed the decrease in cell proliferation, apoptosis inhibition, migration, and invasion and the increase in apoptosis induced by SOX21-AS1 downregulation
To determine whether the SOX21-AS1/miR-24-3p interaction has a role in lung cancer, SOX21-AS1 expression was reduced, and miR-24-3p expression levels were decreased by siRNAs (Figure 3(a,b)). An MTT assay indicated that cell viability was significantly reduced when SOX21-AS1 expression levels decreased, and this was markedly reversed by miR-24-3p depletion (Figure 3(c,d)). Colony formation assays showed similar findings. SOX21-AS1 inhibition decreased cell proliferation, which was abrogated by the miR-24-3p interference (Figure 3(e,f)). In addition, flow cytometric analysis showed that apoptosis was significantly promoted by the silencing of SOX21-AS1, and this was rescued upon inhibition of miR-24-3p (Figure 4). Transwell experiments showed that both migration and invasion abilities were suppressed by SOX21-AS1 knockdown, but miR-24-3p decreased the inhibition of migration and invasion (Figure 5(a,b)). Thus, the SOX21-AS1/miR-24-3p axis increased cell proliferation, inhibited apoptosis, and promoted cell migration and invasion.
Figure 3.
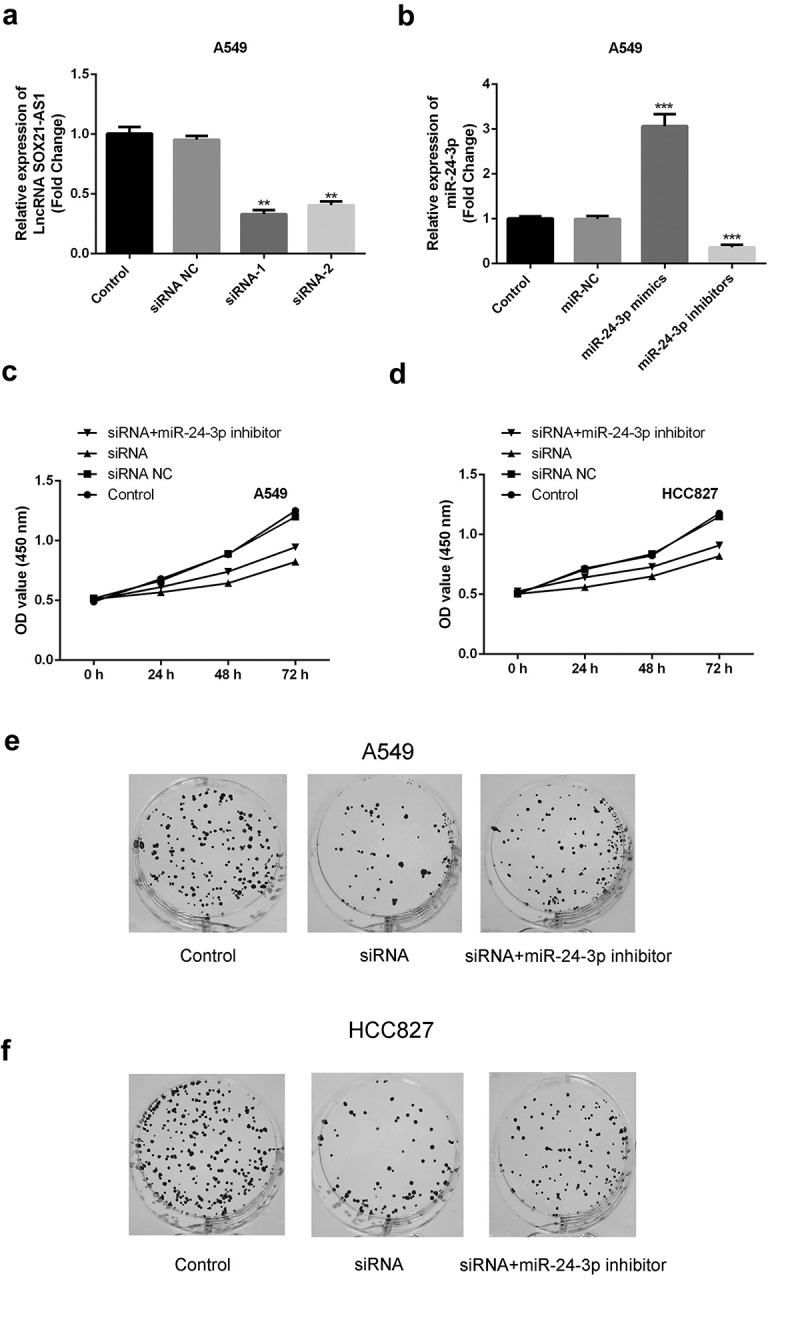
miR-24-3p inhibition reversed the decrease in cell proliferation induced by SOX21-AS1 downregulation
(a) The knockdown efficiency of siRNAs (siRNA-1 and siRNA-2) targeting SOX21-AS1 was examined by RT-qPCR. **P < 0.01 vs control. (b) The expression levels and inference efficacy of miR-24-3p mimics and an miR-24-3p inhibitor were measured by RT-qPCR. ***P < 0.001 vs control. (c-d) An MTT assay was used to evaluate the viability of A549 and HCC827 cells in four different groups. (e-f) Colony formation assays were used to assess the cell proliferation ability of A549 and HCC827 cells.
Figure 4.
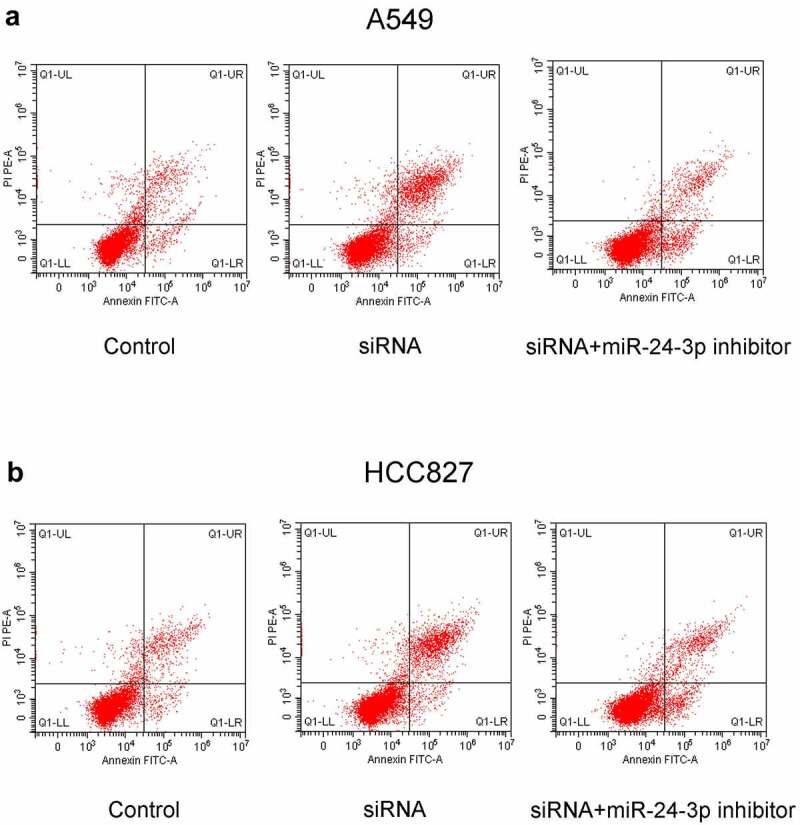
miR-24-3p inhibition decreased apoptosis induced by SOX21-AS1 downregulation
Apoptosis of A549 (a) and HCC827 (b) cells was analyzed by flow cytometry.
Figure 5.
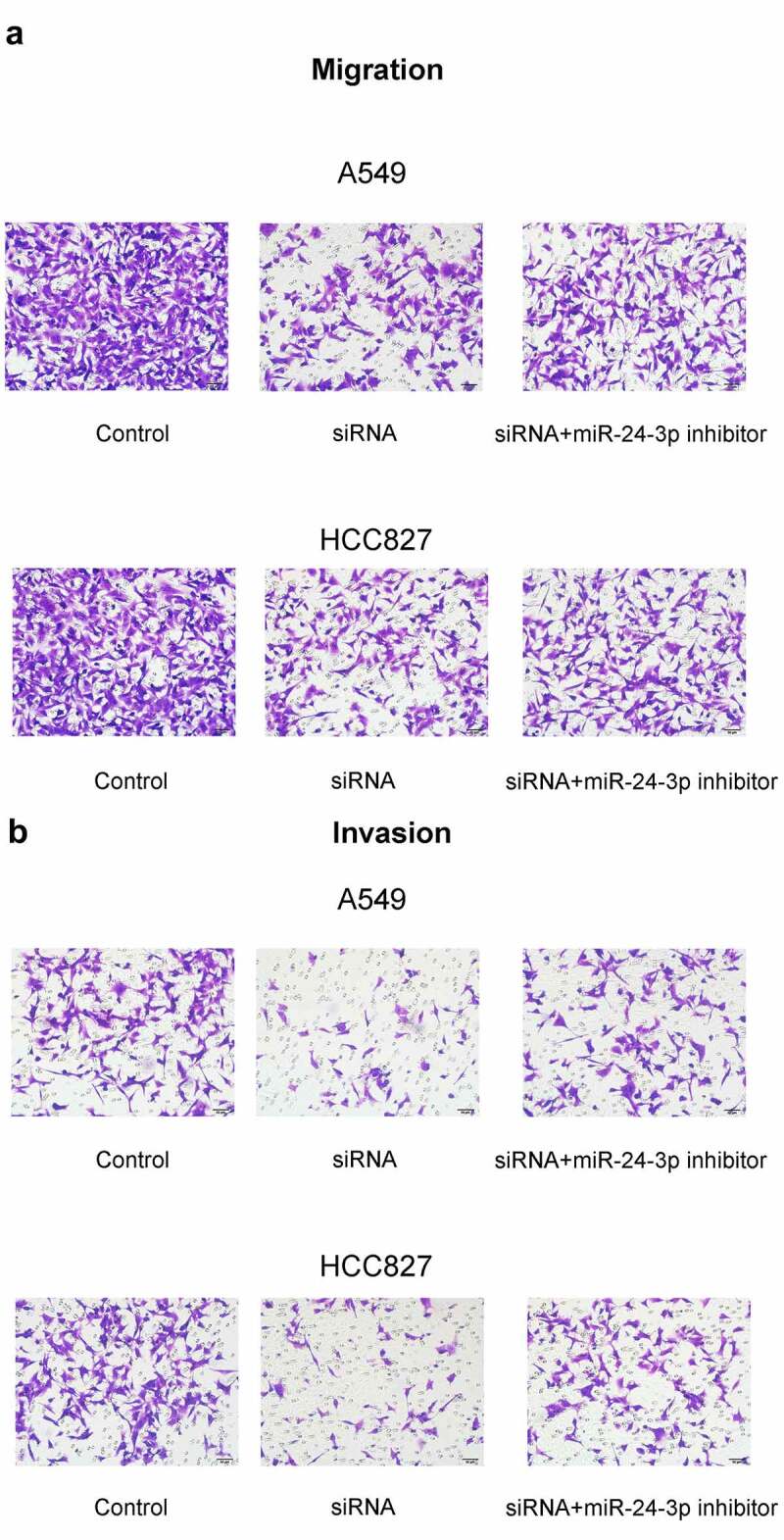
miR-24-3p depletion rescued the decrease in cell migration and invasion induced by SOX21-AS1 downregulation
(a-b) The ability of A549 and HCC827 cells to migrate and invade were assessed via transwell experiments.
miR-24-3p depletion reversed the decrease in tumor formation induced by SOX21-AS1 downregulation
To confirm the function of the SOX21-AS1/miR-24-3p axis in vivo, animal models were established. Tumor volume markedly decreased upon SOX21-AS1 depletion, but significantly increased when miR-24-3p expression levels were reduced (Figure 6(a)). A similar result was observed for tumor size. Tumors in the siRNA treatment group were significantly smaller than those in the control group, but a moderate tumor size was observed in the siRNA+miR-24-3p inhibitor group (Figure 6(b)). Taken together, these results showed that SOX21-AS1 downregulation inhibited tumor formation, and this effect was reversed upon miR-24-3p silencing.
Figure 6.
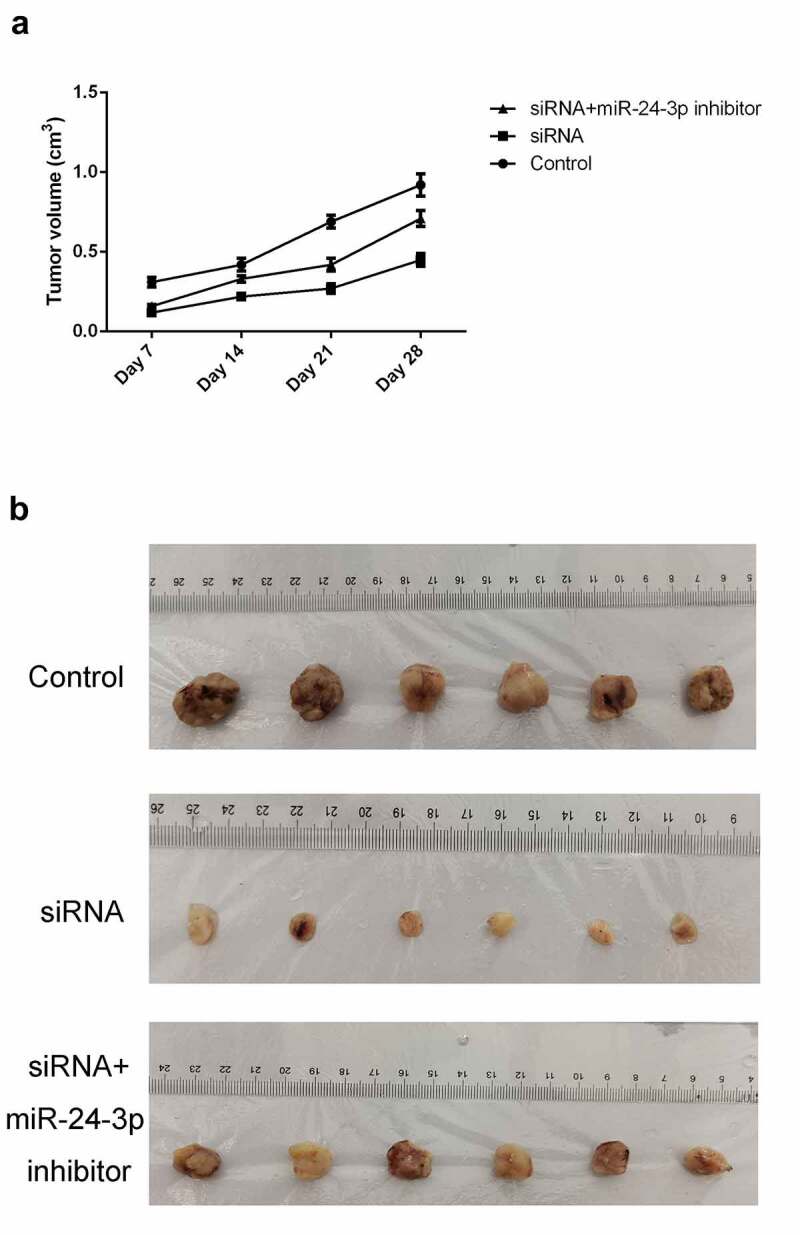
miR-24-3p depletion reversed the decrease in tumor formation induced by SOX21-AS1 downregulation
(a) Tumor volumes of mice in the control, siRNA, and siRNA+miR-24-3p inhibitor groups. (b) Xenografted tumors from mice in the above three groups are shown.
miR-24-3p targeted PIM2, which was overexpressed in lung cancer
The downstream mechanism was further investigated. The predicted binding sequence of miR-24-3p with PIM2 is shown in Figure 7(a). PIM2 levels were markedly higher in lung cancer tissues than in non-tumor tissues (Figure 7(b)). Luciferase reporter assays confirmed that the overexpression of miR-24-3p markedly reduced the luciferase activity of the wild-type PIM2 construct (Figure 7(c)). Moreover, the levels of PIM2 were higher in cancerous cells than in normal BEAS-2B cells (Figure 7(d)).
Figure 7.
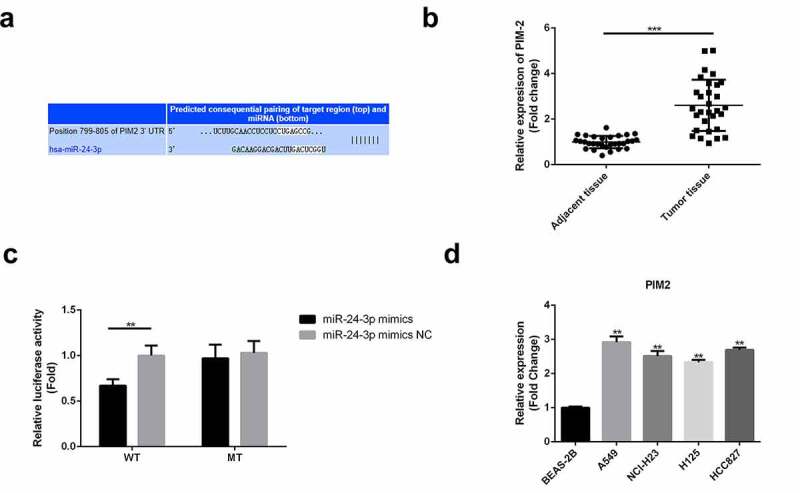
miR-24-3p targeted PIM2, which was overexpressed in lung cancer
(a) The binding sequences of miR-24-3p and PIM2 were predicted using Targetscan. (b) RT-qPCR was used to analyze PIM2 expression in cancer and non-cancer tissues. ***P < 0.001 vs adjacent tissues. (c) Luciferase reporter assays were used to confirm the targeting of PIM2 by miR-24-3p. **P < 0.01 vs miR-24-3p mimic NC. (d) RT-qPCR analysis of PIM2 expression in cancer and normal cells. **P < 0.01 vs BEAS-2B cells.
miR-24-3p over-expression down-regulated the mRNA and protein expression levels of PIM2, inhibited cell proliferation, and promoted the apoptosis
miR-24-3p was overexpressed to explore its impact on PIM2 expression and cellular activity. The mRNA and protein expression levels of PIM2 were downregulated upon overexpression of miR-24-3p (Figure 8(a)). MTT assays showed that cell proliferation was markedly repressed by the overexpression of miR-24-3p (Figure 8(b)). Flow cytometric analysis showed that miR-24-3p overexpression increased apoptosis (Figure 8(c)).
Figure 8.
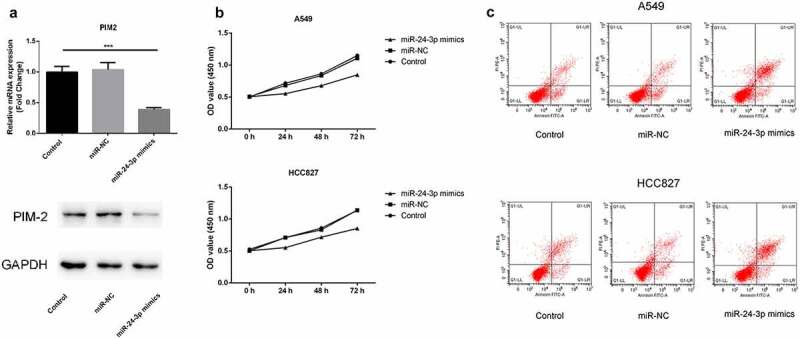
miR-24-3p over-expression down-regulated the mRNA and protein expression levels of PIM2, inhibited proliferation, and promoted apoptosis
(a) RT-qPCR and western blotting were used to measure mRNA and protein expression levels in A549 cells. ***P < 0.001 vs control. (b) MTT experiments were used to analyze the proliferation of the two cell types. (c) Flow cytometry was used to detect the apoptotic ability of the two tumor cell types.
Discussion
The levels of SOX21-AS1 were markedly increased and the levels of miR-24-3p were markedly decreased in cancerous tissues and cells. miR-24-3p was found to interact with SOX21-AS1. The SOX21-AS1/miR-24-3p axis increased the proliferation, migration, and invasion of A549 and HCC827 cells. Animal models further confirmed the tumor-promoting role of the SOX21-AS1/miR-24-3p pathway. PIM2, which showed elevated levels in lung cancer, was the target of miR-24-3p. The targeting of PIM2 by miR-24-3p disrupted the cellular activities of lung cancer cells.
As a newly discovered lncRNA, SOX21-AS1 has been shown to increase tumor progression. SOX21-AS1 facilitates the progression of cervical cancer by targeting the miR-7/VDAC1 pathway [34]. SOX21-AS1 also positively regulates the progression of triple-negative breast cancer via the miR-520a-5p/ORMDL3 axis [35]. SOX21-AS1 increases the proliferation and invasion properties of gliomas by increasing PAK7 levels through the absorption of miR-144-3p [36]. Furthermore, two studies have demonstrated the oncogenic function of SOX21-AS1 in lung cancer. SOX21-AS1 predicts prognosis and potentiates proliferation in lung adenocarcinoma, and its silencing represses migration and invasion by regulating GATA6, which decreases the levels of TSPAN8 [18,19]. In this study, SOX21-AS1 was also highly expressed in both lung cancer tissues and cells, which is consistent with the findings of previous studies. The results in this study and others implied that SOX21-AS1 is a oncogenic lncRNA in lung cancer.
To further elucidate the novel molecular mechanism of SOX21-AS1 in lung cancer, DIANA-LncBase v2 was used to identify the binding sequence between SOX21-AS1 and miR-24-3p. Several studies have demonstrated the role of miR-24-3p as a tumor suppressor. For example, silencing the lncRNA, CCAT1, decreases paclitaxel resistance in prostate cancer by upregulating miR-24-3p and decreasing FSCN1 levels [37,38]. miR-24-3p inhibits the progression of pancreatic ductal adenocarcinoma by decreasing LAMB3 expression levels [39]. In addition, miR-24-3p can inhibit lung adenocarcinoma progression by mediating FGFR3 signaling [23]. In the present study, we observed that miR-24-3p levels were markedly lower in lung cancer tissues and cells than in normal tissues and cells. Moreover, miR-24-3p interacted with SOX21-AS1. Functional assays confirmed that miR-24-3p interference abrogated the inhibition of SOX21-AS1 downregulation on cell proliferation, migration, and invasion, and the promotion of cell apoptosis induced by the downregulation of SOX21-AS1. These results indicated that SOX21-AS1 induces the apoptosis and inhibits the growth of the lung cancer cells via negatively regulating the miR-24-3p expression.
Based on these results, an in-depth investigation of the associated mechanism was performed. Using Targetscan, miRDB, GO, and KEGG, PIM2 was predicted to be the target of miR-24-3p. According to previous reports, PIM2 promotes the progression of a diverse range of cancers. In liver cancer, the inhibition of PIM2 decreases cell proliferation by modulating the cell cycle [24]. PIM2 facilitates HCC progression via NF-κB signaling [39]. PIM2 also interacts with tristetraprolin, thereby promoting breast cancer progression [40]. Consistent with the results from previous studies, lung cancer tissues and cells were found to have relatively high expression levels of PIM2. Further, functional experiments demonstrated that cell proliferation was inhibited and apoptosis was promoted by the upregulation of miR-24-3p. We confirmed that SOX21-AS1 can interact with miR-24-3p through competitive endogenous RNA mechanism, thereby affecting the expression of PIM2 and relieving the lung cancer development.
Conclusion
In conclusion, this study showed that SOX21-AS1 negatively modulated the miR-24-3p/PIM2 axis to potentiate the proliferation, migration, and invasion capabilities of lung cancer cells. This may offer a novel molecular avenue for research into molecular lung cancer therapies.
Highlights
1. SOX21-AS1 is a newly discovered oncogenic lncRNA
2. SOX21-AS1 is up-regulated in lung cancer
3. SOX21-AS1 negatively modulated the miR-24-3p/PIM2 axis
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article.
References
- [1].Chen W, Zheng R, Zhang S, et al. Cancer incidence and mortality in China, 2013. Cancer Lett. 2017;401:63–71. [DOI] [PubMed] [Google Scholar]
- [2].Nasim F, Sabath BF, Eapen GA.. Lung Cancer. Med Clin North Am. 2019;103(3):463–473. [DOI] [PubMed] [Google Scholar]
- [3].Cao M, Chen W.. Epidemiology of lung cancer in China. Thorac Cancer. 2019;10(1):3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hensing T, et al. A personalized treatment for lung cancer: molecular pathways, targeted therapies, and genomic characterization. Adv Exp Med Biol. 2014;799:85–117. [DOI] [PubMed] [Google Scholar]
- [5].Peng W, Wang J, Shan B, et al. Diagnostic and prognostic potential of circulating long non-Coding RNAs in non small cell lung cancer. Cell Physiol Biochem. 2018;49(2):816–827. . [DOI] [PubMed] [Google Scholar]
- [6].Huang Q, Wei H, Wu Z, et al. Preferentially expressed antigen of melanoma prevents lung cancer metastasis. PLoS One. 2016;11(7):e0149640. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wu B, et al. [Comparison of the survival time in the non-small cell lung cancer patients with different organ metastasis]. Zhongguo Fei Ai Za Zhi. 2019;22(2):105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Song Z, Liu Y. [Progress of liquid biopsy in early diagnosis of lung cancer]. Zhongguo Fei Ai Za Zhi. 2018;21(8):620–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang J, Su Z, Lu S, et al. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta. 2018;485:229–233. [DOI] [PubMed] [Google Scholar]
- [10].Li J, Li Z, Zheng W, et al. LncRNA-ATB: an indispensable cancer-related long noncoding RNA. Cell Prolif. 2017;50(6):e12381. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Charles Richard JL, Eichhorn PJA. Platforms for Investigating LncRNA Functions. SLAS Technol. 2018;23(6):493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wei GH, Wang X. lncRNA MEG3 inhibit proliferation and metastasis of gastric cancer via p53 signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21(17):3850–3856. [PubMed] [Google Scholar]
- [13].Deng SJ, Chen H-Y, Ye Z, et al. Hypoxia-induced LncRNA-BX111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene. 2018;37(44):5811–5828. . [DOI] [PubMed] [Google Scholar]
- [14].Zheng L, Xiang C, Li X, et al. STARD13-correlated ceRNA network-directed inhibition on YAP/TAZ activity suppresses stemness of breast cancer via co-regulating Hippo and Rho-GTPase/F-actin signaling. J Hematol Oncol. 2018;11(1):72. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yang CM, Wang T-H, Chen H-C, et al. Aberrant DNA hypermethylation-silenced SOX21-AS1 gene expression and its clinical importance in oral cancer. Clin Epigenetics. 2016;8(1):129. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wei AW, Li LF. Long non-coding RNA SOX21-AS1 sponges miR-145 to promote the tumorigenesis of colorectal cancer by targeting MYO6. Biomed Pharmacother. 2017;96:953–959. [DOI] [PubMed] [Google Scholar]
- [17].Wei C, Wang H, Xu F, et al. LncRNA SOX21-AS1 is associated with progression of hepatocellular carcinoma and predicts prognosis through epigenetically silencing p21. Biomed Pharmacother. 2018;104:137–144. [DOI] [PubMed] [Google Scholar]
- [18].Lu X, Huang C, He X, et al. A novel long Non-Coding RNA, SOX21-AS1, indicates a poor prognosis and promotes lung adenocarcinoma proliferation. Cell Physiol Biochem. 2017;42(5):1857–1869. . [DOI] [PubMed] [Google Scholar]
- [19].Xu Y, Wu H, Wu L, et al. Silencing of long non-coding RNA SOX21-AS1 inhibits lung adenocarcinoma invasion and migration by impairing TSPAN8 via transcription factor GATA6. Int J Biol Macromol. 2020;164:1294–1303. [DOI] [PubMed] [Google Scholar]
- [20].Fabris L, Ceder Y, Chinnaiyan AM, et al. The potential of microRNAs as prostate cancer biomarkers. Eur Urol. 2016;70(2):312–322. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Han X, Li Q, Liu C, et al. Overexpression miR-24-3p repressed Bim expression to confer tamoxifen resistance in breast cancer. J Cell Biochem. 2019;120(8):12966–12976. [DOI] [PubMed] [Google Scholar]
- [22].Yin Y, Zhong J, Li S-W, et al. TRIM11, a direct target of miR-24-3p, promotes cell proliferation and inhibits apoptosis in colon cancer. Oncotarget. 2016;7(52):86755–86765. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jing P, Zhao N, Xie N, et al. miR-24-3p/FGFR3 signaling as a novel axis is involved in Epithelial-Mesenchymal transition and regulates lung adenocarcinoma progression. J Immunol Res. 2018;2018:2834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kronschnabl P, et al. Inhibition of PIM2 in liver cancer decreases tumor cell proliferation in vitro and in vivo primarily through the modulation of cell cycle progression. Int J Oncol. 2020;56(2):448–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Uddin N, Kim R-K, Yoo K-C, et al. Persistent activation of STAT3 by PIM2-driven positive feedback loop for epithelial-mesenchymal transition in breast cancer. Cancer Sci. 2015;106(6):718–725. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Niba ETE, Rochmah MA, Harahap NIF, et al. Spinal muscular atrophy: advanced version of screening system with Real-Time mCOP-PCR and PCR-RFLP for SMN1 deletion. Kobe J Med Sci. 2019;65(2):E49–E53. [PMC free article] [PubMed] [Google Scholar]
- [27].Lei KF, Kao CH, Tsang NM. High throughput and automatic colony formation assay based on impedance measurement technique. Anal Bioanal Chem. 2017;409(12):3271–3277. [DOI] [PubMed] [Google Scholar]
- [28].He X, Dai J, Fan Y, et al. Regulation function of MMP-1 downregulated by siRNA on migration of heat-denatured dermal fibroblasts. Bioengineered. 2017;8(6):686–692. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [29].Omar Zaki SS, Kanesan L, Leong MYD, et al. The influence of serum-supplemented culture media in a transwell migration assay. Cell Biol Int. 2019;43(10):1201–1204. [DOI] [PubMed] [Google Scholar]
- [30].Marshall J. Transwell(®) invasion assays. Methods Mol Biol. 2011;769:97–110. [DOI] [PubMed] [Google Scholar]
- [31].Yao J, Gao RY, Luo MH, et al. Possible role of microRNA miRNA-IL-25 interaction in mice with ulcerative colitis. Bioengineered. 2020;11(1):862–871. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lee NP, Chan CM, Tung LN, et al. Tumor xenograft animal models for esophageal squamous cell carcinoma. J Biomed Sci. 2018;25(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang N, Liu JF. MicroRNA (MiR)-301a-3p regulates the proliferation of esophageal squamous cells via targeting PTEN. Bioengineered. 2020;11(1):972–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhang X, Zhao X, Li Y, et al. Long noncoding RNA SOX21-AS1 promotes cervical cancer progression by competitively sponging miR-7/VDAC1. J Cell Physiol. 2019;234(10):17494–17504. . [DOI] [PubMed] [Google Scholar]
- [35].Liu X, Song J, Kang Y, et al. Long noncoding RNA SOX21-AS1 regulates the progression of triple-negative breast cancer through regulation of miR-520a-5p/ORMDL3 axis. J Cell Biochem. 2020;121(11):4601–4611. . [DOI] [PubMed] [Google Scholar]
- [36].Gai SY, Yuan ZH. Long non-coding RNA SOX21-AS1 promotes cell proliferation and invasion through upregulating PAK7 expression by sponging miR-144-3p in glioma cells. Neoplasma. 2020;67(2):333–343. [DOI] [PubMed] [Google Scholar]
- [37].Li X, Han X, Wei P, et al. Knockdown of lncRNA CCAT1 enhances sensitivity of paclitaxel in prostate cancer via regulating miR-24-3p and FSCN1. Cancer Biol Ther. 2020;21(5):452–462. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Huang W, Gu J, Tao T, et al. MiR-24-3p inhibits the progression of pancreatic ductal adenocarcinoma through LAMB3 downregulation. Front Oncol. 2019;9:1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tang X, Cao T, Zhu Y, et al. PIM2 promotes hepatocellular carcinoma tumorigenesis and progression through activating NF-κB signaling pathway. Cell Death Dis. 2020;11(7):510. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ren C, Yang T, Qiao P, et al. PIM2 interacts with tristetraprolin and promotes breast cancer tumorigenesis. Mol Oncol. 2018;12(5):690–704. . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.


