Abstract
Background
The reliable detection of T cell response to COVID-19 or COVID-19 vaccination is important for individual patient care and for monitoring the immune response e.g. in COVID-19 vaccine trials in a standardized fashion.
Objectives and study design
We used blood samples from health care workers (HCW) with or without history of COVID-19 to define test accuracy of a novel interferon-γ release assay (IGRA). For a real-life performance evaluation, we analysed interferon-γ response to complete COVID-19 vaccination in HCW receiving homologous or heterologous vaccination regimens and in patients receiving immunosuppressive or immune modulating therapies.
Results
The assay had a specificity of 100%. Sensitivity of the IGRA to detect past infection was 72.2% after infection more than 5 months ago and 93.8% after COVID-19 up to 5 months ago. Quantitative results showed significant differences between first and second vaccine dose, but no difference between homologous and heterologous vaccination regimen. Immunocompromised patients often had no immune response or isolated T cell or antibody response to complete vaccination.
Conclusions
The novel IGRA proved to be a highly specific tool to detect SARS-CoV-2 specific T cell response to COVID-19 as well as COVID-19 vaccination, with sensitivity getting lower over time. In perspective, it may serve as a standardized tool in COVID-19 vaccine trials and in clinical care of immunosuppressed patients.
Keywords: COVID-19, Adaptive immune response, IFNγ release assay, T cell response, Test accuracy, Health care workers, Immunosuppressed patients
Graphical abstract
1. Background
The role of T cell immune response in SARS-CoV-2 infection is not entirely understood, but data from animal models show an important role for protection against coronavirus disease 2019 (COVID-19) mediated by CD4+ and CD8+ T cells [1, 2]. Since December 2020, the first vaccines against COVID-19 are available but the quality and duration of the immune response to vaccination remains unclear as of yet. Although numerous SARS-CoV-2 antibody detection assays were introduced with unprecedented speed and are now widely in use, the implementation of T cell assays lagged behind. However, besides analysing the antibody response, it will be of importance to investigate the T cell mediated immune response, e.g. in vaccine trials or in a clinical setting for individual patient care. Therefore, easy to perform, validated, and ideally standardized T cell assays are required [3].
Here we evaluated a novel commercially available IFN-γ release assay (IGRA) to analyse the SARS-CoV-2 specific T cell response. IGRA have revolutionized the diagnosis of tuberculosis [4] and several other pathogens [5, 6]. In experimental settings, in-house SARS-CoV-2 IGRA were introduced recently [7]. The main advantage of this type of assay is the possibility to perform them without special equipment and with very short hands-on time. The IFN-γ results are expressed in international units and are therefore truly quantitative.
2. Objectives
For our test accuracy study, we asked immunocompetent healthcare workers (HCW) at Medical center - University of Freiburg to participate and offered measurement of SARS-CoV-2 antibodies. Recruitment was open to HCW who had experienced mild SARS-CoV-2 infection in 2020 as well as HCW without a history of COVID-19 prior to vaccination. After successful evaluation of the assay, we analysed the COVID-19 vaccine associated T cell response in HCW receiving two different vaccine regimens. Finally, the assay was applied in immunocompromised patients to evaluate the test performance in a real-life setting.
3. Study design
3.1. Study population
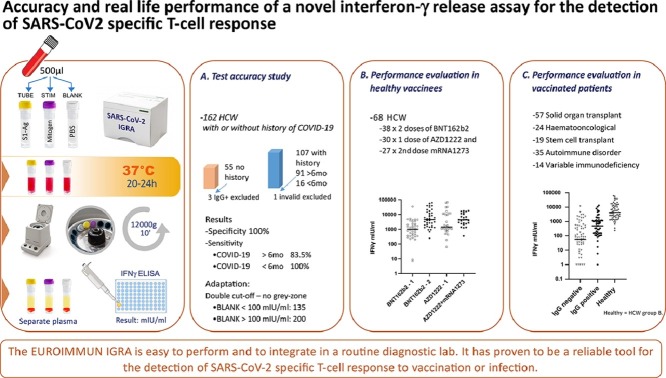
162 HCW agreed to participate at the accuracy study. Of these, 107 had previous SARS-CoV-2 infection (COVID-19 group) proven by a positive SARS-CoV-2 RT-PCR and seroconversion, and 55 were without history of SARS-CoV-2 infection or vaccination (no COVID-19 group). For performance evaluation 68 HCW were tested after vaccination. 38 had received two doses of BNT162b2 (Comirnaty®, BioNTech/Pfizer), 30 had been vaccinated with AZD1222 (Vaxzevria®, AstraZeneca) and of these 27 received a heterologous vaccination scheme with a second dose of mRNA-1273 (Spikevax®, Moderna Biotech). Blood was drawn two to three weeks after the first and second dose. Samples were tested consecutively without knowledge of SARS-CoV-2 antibody status or patient history. For the final real-life performance evaluation, patient samples (n = 149) sent to our routine diagnostic laboratory with the request for SARS-CoV-2 T cell analysis and antibody detection after COVID-19 vaccination were included.
SARS-CoV-2 interferon-γ release assay
The SARS-CoV-2 IGRA (EUROIMMUN, Lübeck, Germany) is based on the SARS-CoV-2 spike protein and was done according to the manufacturer's instructions. For a detailed description of the assay see suppl. 1. IFN-γ concentration was measured using the enzyme-linked immunosorbent assay (ELISA) delivered by the manufacturer in combination with the stimulation tubes. The manufacturer suggested cut-off is at >200 mIU/ml including a grey zone of 100 – 200 mIU/ml.
IGRA Test kits for the first evaluation of the assay were provided by EUROIMMUN free of charge.
3.2. Detection of SARS-CoV-2 antibodies
We used three different SARS-CoV-2 antibody assays to rule out past infection in the no COVID-19 and in the vaccine cohorts: Details on the immunoassays are shown in Supplement 1.
3.3. Statistical analysis
Data were analysed using IBM SPSS Statistics version 27 software, GraphPad Prism version 9, and MedCalc version 19. For detailed description of statistical analysis see supplemental material. For test accuracy calculation, we used two different approaches: we defined grey zone results as either positive or negative [8]. As data had a non-Gaussian distribution, we used nonparametric tests throughout. In order to evaluate the manufacturers’ cut-off definition we performed receiver operating characteristic (ROC) analysis to define an optimum cut-off value at the Youden maximum index value.
4. Ethics
The study was approved by the ethics committee of Albert-Ludwigs University Freiburg (#20–1271, Nov 24, 2020 and #20–1271_1, Jan 18, 2021). Written informed consent was obtained from all participants.
5. Resuts
We enroled 162 HCW for the validation study. All HCW in the COVID-19 group had referred mild symptoms. Baseline characteristics of the HCW study groups are shown in Table 1 .
Table 1.
Baseline characteristics of the 162 health care workers.
| COVID-19 ≥ 6 months ago, n = 91 | COVID-19 <6 months ago, n = 16 | No COVID-19 group n = 55 | |
|---|---|---|---|
| Age in years, mean (range) | 43.9 (22 – 64) | 38.1 (22 – 60) | 42.58 (20 – 70) |
| Sex | |||
| Female% | 64.8 | 50 | 67.3 |
| Male% | 35.2 | 50 | 32.7 |
5.1. IFN-γ concentrations in the no COVID-19 group
Out of 55 samples from the no COVID-19 group, 50 had IFN-γ concentrations below 100 mIU/ml, two between 100 and 200 mIU/ml, and three above 200 mIU/ml. Serum samples of these three individuals were reactive in several antibody assays suggesting past infection. Thus, we excluded these samples from further analysis. Serum samples from the 52 remaining individuals of this cohort tested negative with three different SARS-CoV-2 antibody assays and were therefore defined as truly negative. Specificity of the assay using 200 mIU/ml and 100 mIU/ml was 100% [95% confidence interval (CI) 93.2%−100%] and 96.2% (95% CI 86.8%−99.5%), respectively.
5.2. IFN-γ concentrations in HCW with previous SARS-CoV-2 infection (COVID-19 group)
A total of 107 samples from HCW with previous SARS-CoV-2 infection were available to us. In one case, the assay was invalid, the sample was thus excluded from further analysis. 80 of the remaining 106 HCW (75.4%) yielded IFN-γ concentrations above 200 mIU/ml, 15 (14.2%) between 100 and 200 mIU/ml and 11 (10.3%) below 100 mIU/ml. Overall sensitivity to detect past infection was 75.4% using 200 mIU/ml as cut-off and 89.7% defining grey-zone results as positive. Of the 106 HCW, 90 had blood samples drawn more than six months after RT-PCR proven SARS-CoV-2 infection. Eleven (12.2%) yielded IFN-γ concentrations below 100 mIU/ml, 14 (15.6%) between 100 and 200 mIU/ml and 65 (72.2%) above 200 mIU/ml. In this subgroup, we calculated a sensitivity of 72.2% defining IFN-γ concentrations of 200 mIU/ml as true positive and 87.8% with grey-zone results defined as positive. Another 16 individuals were infected two to five months ago, 15 of which had IFN-γ concentrations above 200 mIU/ml and one was between 100 and 200 mIU/ml. Sensitivities for recent infection in this subgroup were 93.8% or 100% defining grey-zone results as negative or positive, respectively.
5.3. ROC analysis, background IFN-γ concentrations and adapted performance calculation
Performing ROC analysis with the “no COVID-19 group” as true negative and the “COVID-19 group” as true positive cohort, highest Youden index was seen at 135 mIU/ml with a specificity of 98.1% (95% CI 89.8%−99.9%) and a sensitivity of 93.4% (95% CI 89.2–96.3%). Background IFN-γ stimulation (BLANK values) was low in most participants (mean 35.2 mIU/ml, range 0–1520 mIU/ml), but 22/158 (13.9%) showed elevated background stimulation with IFN-γ concentrations above 100 mIU/ml including the two false positive samples (see above). We therefore aimed to include the BLANK values into the result interpretation and to circumvent the need of a grey-zone. Subsequently, we used a cut-off of equal or above 135 mIU/ml in individuals with background stimulation below 100 mIU/ml and a cut-off of 200 mIU/ml in individuals with higher background stimulation. Using our BLANK-value adapted approach specificity reached 100% and overall sensitivity to detect past infection was 86.8% (100% for infection less than six months and 83.5% for infection more than 5 months ago). Predictive values and accuracy of the different cut-off strategies are shown in Table 2 .
Table 2.
Diagnostic test accuracy of the EUROIMMUN interferon-γ release assay using different cut-offs at an estimated COVID-19 disease prevalence of 10%.
| Cut-off >100 mIU/ml | Cut-off ≥200 mIU/ml | Adapted cut-off* | |
|---|---|---|---|
| Sensitivity% (95% CI) | 89.6 (82.2 – 94.7) | 72.2 (61.8 – 81.2) | 86.8 (78.8 – 92.6) |
| Specificity | 96.2 (86.8 – 99.5) | 100.0 (93,2.7 – 100,0) | 100 (93.2 – 100,0) |
| Negative predictive value | 98.8 (97.9 – 99.3) | 97.1 (95.9 – 97.8) | 98.6 (97.7 – 99.1) |
| Positive predictive value | 72.2 (39.9 – 90.9) | 100,0 | 100,0 |
| Accuracy | 95.5 (90.9 – 98.2) | 97.2 (93.0 – 99.2) | 98.7 (95.4 – 99.8) |
Adapted cut-off: Cut-off ≥135 mIU/ml if BLANK < 100 mIU/ml and ≥200 mIU/ml if BLANK ≥100 mIU/ml mIU/ml.
Of note, 20 out of 106 (18.9%) HCW several months after SARS-CoV-2 infection were negative for SARS-CoV-2 S1-IgG but tested positive for IFN-γ stimulation using the BLANK value adapted cut-off. Three of five HCW who were non-reactive even in the highly sensitive total-antibody assays were positive showing IFN-γ concentrations between 136 and 630 mIU/ml (Supplemental Table 2).
5.4. Real life performance: IFN-γ concentrations in HCW after COVID-19 vaccination
Next, we analysed IFN-γ concentrations in HCW after COVID-19 vaccination (Table 3 ).
Table 3.
Baseline characteristics of 68 health care workers after COVID-19 vaccination.
| Vaccine group BNT162b2, n = 38 | Vaccine group AZD1222, n = 30 | Vaccine group AZD1222/mRNA-1273, n = 27 | |
|---|---|---|---|
| Age in years, mean (range) | 42.6 (19 - 63) | 44.2 (23 - 64) | 44.9 (23 - 64) |
| Sex | |||
| Female% | 76.3 | 88.9 | 88.9 |
| Male% | 23.7 | 11.1 | 11.1 |
In the BNT162b2 group, 36/38 (95%) had IFN-γ concentrations above 200 mIU/ml after the first dose and 38/38 (100%) after the second dose, respectively. In individuals receiving AZD1222, 28/30 (93%) had IFN-γ concentrations above 200 after the first dose, one had 190 mIU/ml (positive using the adapted cut-off definition) and one did not show specific IFN-γ release (64 mIU/ml). All 27 HCW who were vaccinated with the Moderna vaccine mRNA1273 after receiving a first dose with AZD1222 had IFN-γ concentrations clearly above 200 mIU/ml.
5.5. Comparison of IFN-γ concentrations in COVID-19 patients versus vaccinated HCW
Median IFN-γ concentration was significantly different between individuals with past infection more than 6 months vs. less than 6 months ago (Mann-Whitney U test, p = 0.001) (Fig. 1 ). IFN-γ concentrations after first dose of BNT162b2 vaccine were similar as those after recent infection and significantly lower than concentrations after the second dose (Wilcoxon test, p = 0.001). Median IFN-γ concentration after the first dose of AZD1222 was higher than after the first dose of BNT162B2 with a large range (Mann-Whitney U test, p = 0.04). Median concentration after heterologous booster immunization with mRNA1273 was similar to median concentration after second dose with BNT162b2. Mean, median, standard deviation and range are shown in Table 4 .
Fig. 1.
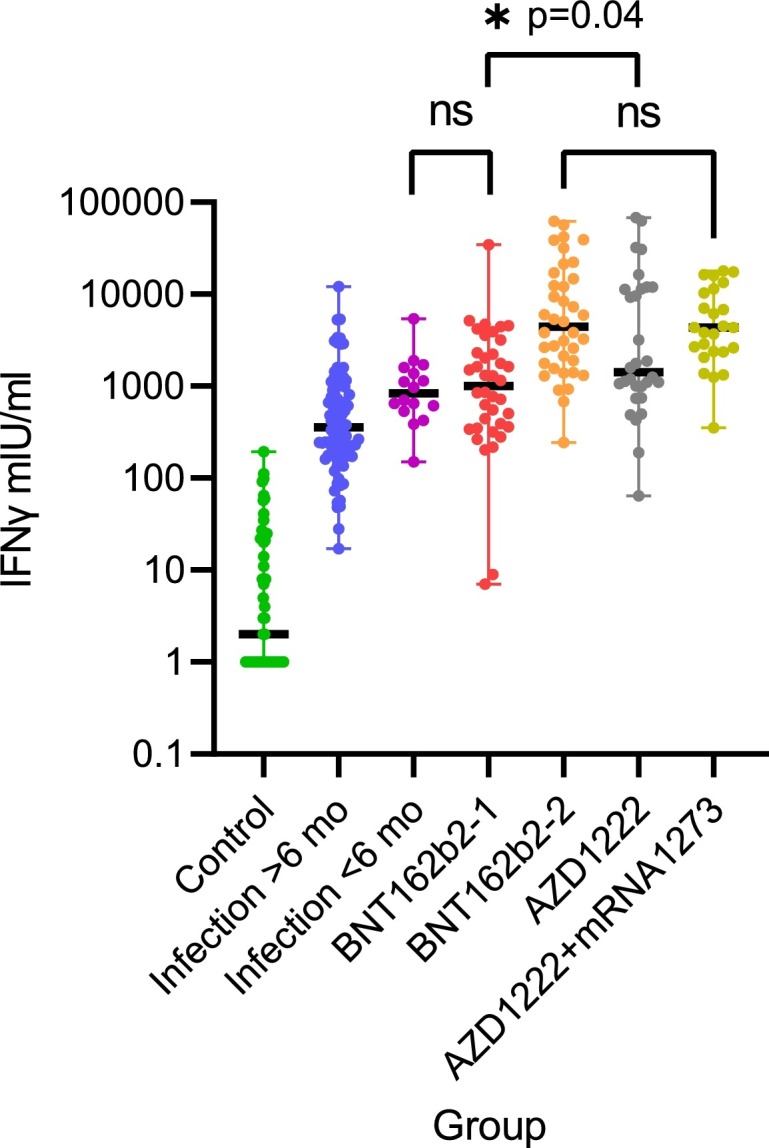
IFN-y release after stimulation with SARS-CoV-2 spike antigen in different groups. Differences are highly significant with p < 0.001 unless indicated. BNT162b2–1 and −2 denotes health care workers (HCW) after the first and second dose of BNT162b2 (n = 38), AZD1222 denotes HCW after first dose of AZD1222 (n = 30) and AZD1222+mRNA1273 after the second dose with mRNA-1273 (heterologous vaccination scheme, n = 27). ns = not significant.
Table 4.
Interferon-γ results in mIU/ml in the different study groups.
| Study group | n | Mean | SD* | Median | Min | Max |
|---|---|---|---|---|---|---|
| No COVID-19 | 52 | 20 | 38 | 2 | 0 | 193 |
| Past infection >6mo | 90 | 885 | 1584 | 356 | 17 | 12,149 |
| Past infection <6mo | 16 | 1206 | 1224 | 838 | 151 | 5383 |
| BNT162b2 1. dose | 38 | 2445 | 5522 | 1008 | 7 | 34,300 |
| BNT162b2 2. dose | 38 | 14,269 | 18,437 | 5198 | 245 | 67,049 |
| AZD1222 1. dose | 30 | 9847 | 17,207 | 1421 | 64 | 67,748 |
| AZD1222/mRNA1273 | 27 | 6492 | 16,969 | 4440 | 245 | 67,049 |
| Immunocompromised patients, IgG positive | 54 | 1475 | 2852 | 501 | 0 | 13,250 |
| Immunocompromised patients, IgG negative | 76 | 583 | 1553 | 59 | 0 | 11,700 |
SD Standard deviation.
5.6. Real life performance: IFN-y response in immunocompromised patients
After validation and implementation of the assay in our routine diagnostic lab, we received test orders for analysing antibody and T-cell immune response of several immunocompromised patients after complete COVID-19 vaccination. These patients (n = 149) had various underlying diseases and received immunosuppressive or immune modulating therapies or had variable immunodeficiency (Table 5 ). The majority of immunocompromised patients showed an inadequate immune response to vaccination.
Table 5.
Combined immune response (detection of SARS-CoV-2 S1-IgG and/or IFN-y response) to vaccination in immunocompromised patients and healthy adults.
| Underlying disease | S1-IgG negative AND IGRA* negative or invalidn(%) | IGRA* positive, S1-IgG negativen(%) | S1-IgG positive, IGRA* negative or invalidn(%) | IGRA* and S1-IgG positiven(%) | IGRA* invalidn(%) |
|---|---|---|---|---|---|
| Lung transplant (n = 33) | 29 (87,9) | 1 (3.0) | 2 (6.0) | 1 (3.0) | 8 (24.2) |
| Kidney transplant (n = 24) | 14 (58.3) | 3 (12.5) | 5 (20.8) | 2 (8.3) | 9 (37.5) |
| Haemato-oncological (n = 24) | 12 (50) | 6 (25.0) | 2 (8.3) | 4 (16.6) | 3 (12.5) |
| Stem cell transplant (n = 19) | 3 (15.8) | 3 (15.8) | 6 (31.6) | 7 (36.8) | 3 (15.8) |
| Autoimmune disorder (n = 35) | 7 (20.0) | 14 (40.0) | 4 (11.4) | 10 (28.6) | 2 (5.7) |
| Variable immunodeficiency (n = 14) | 6 (42.9) | 1 (7.1) | 1 (7.1) | 6 (42.8) | 0 |
| HCW BNT162b2 (n = 38) | 0 | 0 | 0 | 38 (100) | 0 |
| HCW AZD1222/mRNA1273 (n = 27) | 0 | 0 | 0 | 27 (100) | 0 |
IGRA = Interferon Gamma Release Assay.
6. Discussion
In this study, the EUROIMMUN SARS-CoV-2 IGRA demonstrated a cut-off dependant specificity of 96.3–100% and sensitivity of 75.4–86.9% to detect past infection. We used a rigorously tested set of samples to evaluate the IGRA thoroughly. The assay proved to be a sensitive and specific tool for detecting the cellular immune response to COVID-19 or COVID-19 vaccination. Technically, it is easy to perform and can be rapidly implemented in routine diagnostic laboratories. This will assist e.g. in patient care and future COVID-19 vaccination trials as it will allow on site testing of the cellular immune response using standardized and widely available assays.
Understanding the immune response to COVID-19 is essential for the development of preventive strategies against COVID-19. Since the end of 2020 several vaccines have been authorized by medicines agencies worldwide and mass vaccination campaigns in some countries have been very successful in reducing case numbers significantly [9, 10]. Nevertheless, breakthrough infections occur with all vaccines and it will be important to define the correlates of protection after vaccination as well as after infection [11], [12], [13], [14], [15]. From recent communications of breakthrough infections in healthy and immunosuppressed adults it appears that humoral response alone may not be sufficient for protection against infection and disease [12, 15, 16]. Longitudinal studies of COVID-19 patients have observed a significant association of a specific T cell response with milder disease in the absence of SARS-CoV-2 IgG seroconversion and even in agammaglobulinaemic and B-cell depleted patients suggesting that T cell responses may be important for control of SARS-CoV-2 infection [17], [18], [19], [20], [21], [22], [23]. Of note, T cell responses in convalescents and after COVID-19 vaccination seem not to be affected by mutations found in SARS-CoV-2 variants of concern [24], [25], [26], [27], whereas neutralizing capacity of antibodies is significantly reduced [28]. It is therefore of importance to measure T cell mediated reactivity to evaluate immune response to vaccination.
For multicentre vaccination studies standardized and easy to perform assays to measure T cell response in large scale manner are required. IFN-γ is a key cytokine for T cell mediated immune response to specific antigens [29, 30]. The expression of IFN-γ can therefore be used as a marker of pathogen-specific immunity. Of note, the results of different assay types for IFN-γ release upon stimulation are difficult to compare [29] and there is no true gold standard for the measurement of cellular immune response. Thus, to evaluate the specificity of the assay other information has to be used [31]. We used samples from HCW without history of COVID-19 or COVID-19 vaccination and without detectable SARS-CoV-2 antibodies for specificity analysis. Several authors have observed SARS-CoV-2 specific T cells in negative cohorts [32, 33] even using spike protein peptide pools for stimulation similar to the here described assay. Using the adapted cut-off strategy the IGRA was negative in all 52 healthy controls in our study, showing very high specificity, which is an essential requirement for its suitability for large-scale studies. It further supports the notion that the assay does not interfere with pre-existing and cross-reactive T cells from previous common/seasonal coronavirus infections.
Diagnosis of past infection without history of PCR positivity is difficult using SARS-CoV-2 antibody-based testing alone, as antibodies wane after several months and in some cases become undetectable even with the most sensitive antibody assays [34]. Highly specific T cell assays may help proving specificity of low positive or grey zone antibody results and can also be used to confirm past infection in long-covid patients without PCR results.
For the use in vaccination studies quantification of the T cell response must be reliable and reproducible. We were able to show significant differences between the first and the second vaccination amongst groups as well as individuals and between different vaccines. Similar results have been reported recently in a study using the same IGRA [35]. Reproducibility was good and the assay showed high tolerance against variations of standard procedures (supplemental material). These properties make the assay highly suitable for routine diagnostic laboratories and opens up possibilities to analyse the immune response beyond using antibody detection assays in vaccinated patients especially under immunosuppressing or immune modulating medications. Importantly, we were able to show that B-cell depleted patients in most cases mount a strong T cell response to COVID-19 vaccination and are thus probably protected against severe disease [23]. However, this was not seen in most patients after organ transplantation confirming previous studies [36]. These patients can only be protected by vaccination of their close contacts and non-pharmaceutical intervention strategies.
Limitation for the use of such a biological assay is the functionality of the T cell response. Even low-dose steroid treatment may interfere with IGRA testing, as has been shown for Quantiferon TB in immunocompetent children (34). Further, the interpretation of grey zone results is always difficult, so we developed a double cut-off strategy including BLANK results in the cut-off definition. Diagnostic test accuracy analysis showed best accuracy with similar negative predictive values using our adapted cut-off strategy. However, the adapted cut-off should be validated independently in other cohorts.
In conclusion, the EUROIMMUN IGRA is an easy to perform assay for the detection of SARS-CoV-2 specific T cell response with high sensitivity and specificity. During a performance evaluation of the assay we were able to show that patients on immunosuppressive regimens may have isolated T cell or antibody response or in several cases do not respond at all. We propose that measurement of immune response to vaccination in immunocompromised patients should always include an analysis of T cell response and the EUROIMMUN IGRA proved to be highly suitable for this purpose.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Statement of meetings where the information has been previously presented
This work has not been presented at any meetings.
CREdiT author statement
Daniela Huzly, Marcus Panning: Conceptualization, Methodology, Software, Validation, Writing- Original draft preparation. Franziska Smely: Investigation, Data curation, Visualization. Martin Enders: Writing- Reviewing and Editing, Investigation. Johanna Komp: Data curation, Investigation. Valeria Falcone: Data curation, Software, Formal analysis. Daniel Steinmann: Resources, Writing- Reviewing and Editing, Project Administration.
Declarations of Competing Interest
All authors have nothing to declare.
Acknowledgement
We are grateful to Ingeborg Hanselmann for expert technical assistance. We would like to thank all health care workers and patients for their participation and support.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2022.105098.
Appendix. Supplementary materials
References
- 1.McMahan K., Yu J., Mercado N.B., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertoletti A., Tan A.T., Le Bert N. The T cell response to SARS-CoV-2: kinetic and quantitative aspects and the case for their protective role. Oxford Open Immunol. 2021 doi: 10.1093/oxfimm/iqab006. iqab006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hellerstein M. What are the roles of antibodies versus a durable, high quality T-cell response in protective immunity against SARS-CoV-2? Vaccine. 2020;6 doi: 10.1016/j.jvacx.2020.100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sester M., Sotgiu G., Lange C., et al. Interferon-γ release assays for the diagnosis of active tuberculosis: a systematic review and meta-analysis. Eur. Respir. J. 2011;37:100–111. doi: 10.1183/09031936.00114810. [DOI] [PubMed] [Google Scholar]
- 5.Paouri B., Soldatou A., Petrakou E., et al. Quantiferon-Cytomegalovirus assay: a potentially useful tool in the evaluation of CMV-specific CD8+ T-cell reconstitution in pediatric hematopoietic stem cell transplant patients. Pediatr. Transplant. 2018;22:e13220. doi: 10.1111/petr.13220. [DOI] [PubMed] [Google Scholar]
- 6.Terada K., Itoh Y., Fujii A., Kitagawa S., Ogita S., Ouchi K. Varicella-zoster virus-specific, cell-mediated immunity with interferon-gamma release assay after vaccination of college students with no or intermediate IgG antibody response. J. Med. Virol. 2015;87:350–356. doi: 10.1002/jmv.24031. [DOI] [PubMed] [Google Scholar]
- 7.Murugesan K., Jagannathan P., Pham T.D., et al. Interferon-gamma release assay for accurate detection of SARS-CoV-2 T cell response. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bossuyt P.M., Reitsma J.B., Bruns D.E., et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ. 2015;351:h5527. doi: 10.1136/bmj.h5527. (Clinical research ed) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haas E.J., Angulo F.J., McLaughlin J.M., et al. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS-CoV-2 infections and COVID-19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: an observational study using national surveillance data. Lancet. 2021;397:1819–1829. doi: 10.1016/S0140-6736(21)00947-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez Bernal J., Andrews N., Gower C., et al. Effectiveness of the Pfizer-BioNTech and Oxford-AstraZeneca vaccines on covid-19 related symptoms, hospital admissions, and mortality in older adults in England: test negative case-control study. BMJ. 2021;373:n1088. doi: 10.1136/bmj.n1088. (Clinical research ed) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyagi K., Ghosh A., Nair D., et al. Breakthrough COVID19 infections after vaccinations in healthcare and other workers in a chronic care medical facility in New Delhi, India. Diabetes Metab. Syndr. 2021;15:1007–1008. doi: 10.1016/j.dsx.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CDC. https://www.cdc.gov/vaccines/covid-19/health-departments/breakthrough-cases.html. Available at: https://www.cdc.gov/vaccines/covid-19/health-departments/breakthrough-cases.html 2020.
- 13.Tyagi K., Ghosh A., Nair D., et al. Breakthrough COVID19 infections after vaccinations in healthcare and other workers in a chronic care medical facility in New Delhi, India. Diabetes Metab. Syndr. 2021;15:1007–1008. doi: 10.1016/j.dsx.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teran R.A., Walblay K.A., Shane E.L., et al. Postvaccination SARS-CoV-2 infections among skilled nursing facility residents and staff members - Chicago, Illinois, December 2020-March 2021. Am. J. Transplant. 2021;21:2290–2297. doi: 10.1111/ajt.16634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hacisuleyman E., Hale C., Saito Y., et al. Vaccine breakthrough infections with SARS-CoV-2 variants. N. Engl. J. Med. 2021;384:2212–2218. doi: 10.1056/NEJMoa2105000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song C.C., Christensen J., Kumar D., Vissichelli N., Morales M., Gupta G. Early experience with SARs-CoV-2 mRNA vaccine breakthrough among kidney transplant recipients. Transpl. Infect. Dis. 2021:e13654. doi: 10.1111/tid.13654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarzkopf S., Krawczyk A., Knop D., et al. Cellular immunity in COVID-19 convalescents with PCR-confirmed infection but with undetectable SARS-CoV-2-specific IgG. Emerg Infect Dis. 2021:27. doi: 10.3201/2701.203772. [DOI] [PubMed] [Google Scholar]
- 18.Reynolds C.J., Swadling L., Gibbons J.M., et al. Discordant neutralizing antibody and T cell responses in asymptomatic and mild SARS-CoV-2 infection. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abf3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.19 Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012. doi: 10.1016/j.cell.2020.09.038. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soresina A., Moratto D., Chiarini M., et al. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr. Allergy Immunol. 2020;31:565–569. doi: 10.1111/pai.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montero-Escribano P., Matías-Guiu J., Gómez-Iglesias P., Porta-Etessam J., Pytel V., Matias-Guiu J.A. Anti-CD20 and COVID-19 in multiple sclerosis and related disorders: a case series of 60 patients from Madrid, Spain. Mult. Scler. Relat. Disord. 2020;42 doi: 10.1016/j.msard.2020.102185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bange E.M., Han N.A., Wileyto P., et al. CD8+ T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat. Med. 2021 doi: 10.1038/s41591-021-01386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Redd A.D., Nardin A., Kared H., et al. CD8+ T cell responses in COVID-19 convalescent individuals target conserved epitopes from multiple prominent SARS-CoV-2 circulating variants. Open Forum Infect. Dis. 2021:ofab143. doi: 10.1093/ofid/ofab143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarke A., Sidney J., Methot N., et al. Negligible impact of SARS-CoV-2 variants on CD4 (+) and CD8 (+) T cell reactivity in COVID-19 exposed donors and vaccinees. bioRxiv. 2021 [Google Scholar]
- 26.Woldemeskel B.A., Garliss C.C., Blankson J.N. SARS-CoV-2 mRNA vaccines induce broad CD4+ T cell responses that recognize SARS-CoV-2 variants and HCoV-NL63. J. Clin. Invest. 2021:131. doi: 10.1172/JCI149335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geers D., Shamier M.C., Bogers S., et al. SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccinees. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abj1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z., Schmidt F., Weisblum Y., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagen J., Zimmerman R., Goetz C., et al. Comparative multi-donor study of IFNγ secretion and expression by human PBMCs using ELISPOT side-by-side with ELISA and flow cytometry assays. Cells. 2015;4:84–95. doi: 10.3390/cells4010084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sahin U., Muik A., Derhovanessian E., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 31.Rutjes A.W., Reitsma J.B., Coomarasamy A., Khan K.S., Bossuyt P.M. Evaluation of diagnostic tests when there is no gold standard. A review of methods. Health Technol. Assess. 2007;11 doi: 10.3310/hta11500. iii, ix-51. [DOI] [PubMed] [Google Scholar]
- 32.Grifoni A., Weiskopf D., Ramirez S.I., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braun J., Loyal L., Frentsch M., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587 doi: 10.1038/s41586-020-2598-9. 270-4. [DOI] [PubMed] [Google Scholar]
- 34.Aubry A., Demey B., François C., et al. Longitudinal analysis and comparison of six serological assays up to eight months post-COVID-19 diagnosis. J. Clin. Med. 2021;10 doi: 10.3390/jcm10091815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hillus D., Schwarz T., Tober-Lau P., et al. Safety, reactogenicity, and immunogenicity of homologous and heterologous prime-boost immunisation with ChAdOx1-nCoV19 and BNT162b2: a prospective cohort study. medRxiv. 2021 doi: 10.1016/S2213-2600(21)00357-X. 2021.05.19.21257334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sattler A., Schrezenmeier E., Weber U., et al. Impaired humoral and cellular immunity after SARS-CoV2 BNT162b2 (Tozinameran) prime-boost vaccination in kidney transplant recipients. medRxiv. 2021 doi: 10.1172/JCI150175. 2021.04.06.21254963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.