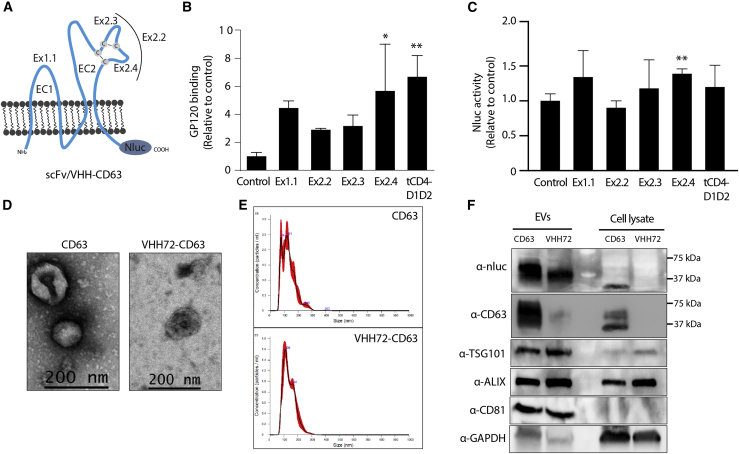
Figure 1.
Characterization of targeted CD63 EVs
(A) Schematic of the CD63 receptor and the insertion sites of the N6 scFv or VHH72 nanobody (Ex1.1, Ex2.2, Ex2.3, or Ex2.4) or a truncated CD4 domains 1 and 2 attached to the N terminus of CD63 (tCD4-D1D2). EC1 and EC2 denote the two main loops of CD63, and cysteine disulfide bonds are highlighted. A Nluc was fused in-frame to the C -terminus. (B) The N6-CD63 EVs were bound to beads and then incubated with gp120, and binding was assessed by flow cytometry, made relative to the CD63 control set at 100%. Error bars represent standard deviation generated from samples treated in triplicate. The p values were generated using a one-way ANOVA compared with the control (∗p < 0.05, ∗∗p < 0.01). (C) HEK293 cells that stably express gp160 were treated with N6-CD63 EVs, and the levels of Nluc were assessed at 18 h post-addition. The Nluc levels were normalized to HEK293 WT cells and made relative to the CD63 control set at 100%. Error bars represent standard deviation generated from samples treated in triplicate. The p values were generated using an unpaired Student’s t test compared with the control (∗∗p < 0.01). (D) TEM and (E) NTA analysis for the CD63 control and VHH72-CD63 EVs. (F) EVs and cell lysates were assessed by western blot for known EV markers (TSG101, ALIX, CD81) and the components of the CD63 fusion protein (Nluc and CD63). GAPDH was included as a loading control. Ladder molecular weights are indicated.