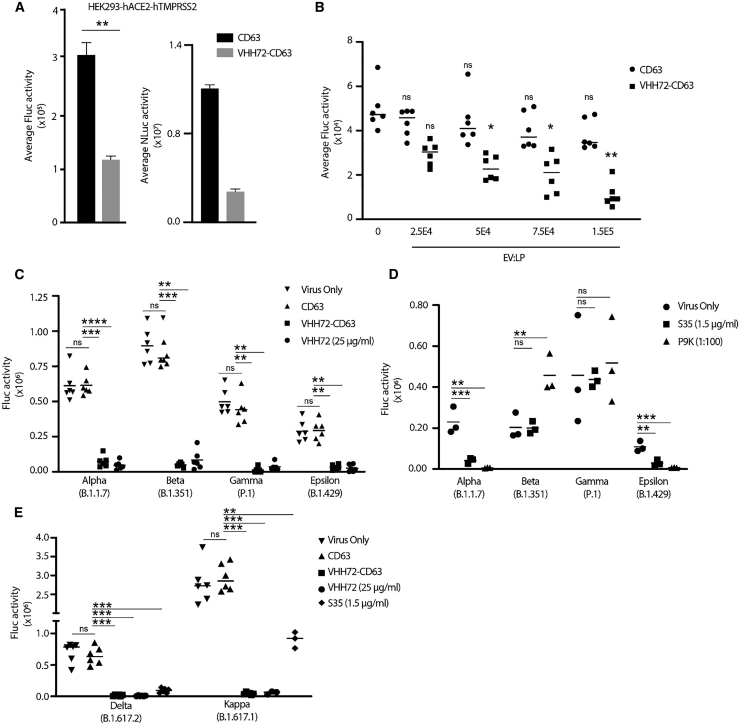
Figure 3.
VHH72-CD63 EVs broadly neutralize SARS-CoV-2 pseudovirus
(A) The pseudotyped spike D614G-R682Q lentiviral particles were incubated with EVs and then transduced on HEK293-hACE2-hTMPRSS2 cells, and the levels of Fluc and Nluc were assessed at 72 h post-transduction. Error bars represent standard deviation generated from samples treated in triplicate. The p values were generated using an unpaired Student’s t -test compared with the CD63 control treated samples (∗∗p < 0.01). (B) The EVs were incubated at increasing concentrations with a set number of lentiviral particles (LP), and transductions were performed as described. Error bars represent standard deviation generated from samples treated in triplicate from two independent experiments. The p values were generated using a one-way ANOVA by comparing the means of the two experiments relative to the virus-only control (∗p < 0.05, ∗∗p < 0.01). (C) The pseudotyped lentiviral particles with the spike protein from the Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), and Epsilon (B.1.429) variants were incubated with 2E9 EVs, and then HEK293-hACE2-hTMPRSS2 cells were transduced. The levels of Fluc were assessed at 48 h post-transduction. The line represents the mean from samples treated in triplicate from two independent experiments. (D) The lentiviral particles pseudotyped with spikes from SARS-CoV-2 VOC were incubated with the S35 mAb (1.5 μg/mL) or CCP (P9K; 1:100 dilution), and then HEK293-hACE2-hTMPRSS2 cells were transduced. The line represents the mean from an experiment performed in triplicate. (E) VHH72-CD63 EVs were tested against lentivirus pseudotyped with the Delta (B.1.617.2) and Kappa (B.1.617.1) spike variants. The line represents the mean from samples treated in triplicate from two independent experiments, except for the S35- and recombinant VHH72-treated samples for the Kappa variant, which were generated from one independent experiment. The p values for (C), (D), and (E) were generated using a one-way ANOVA by comparing the means of the two experiments relative to the CD63 (C and E) or virus-only control (D) (∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).