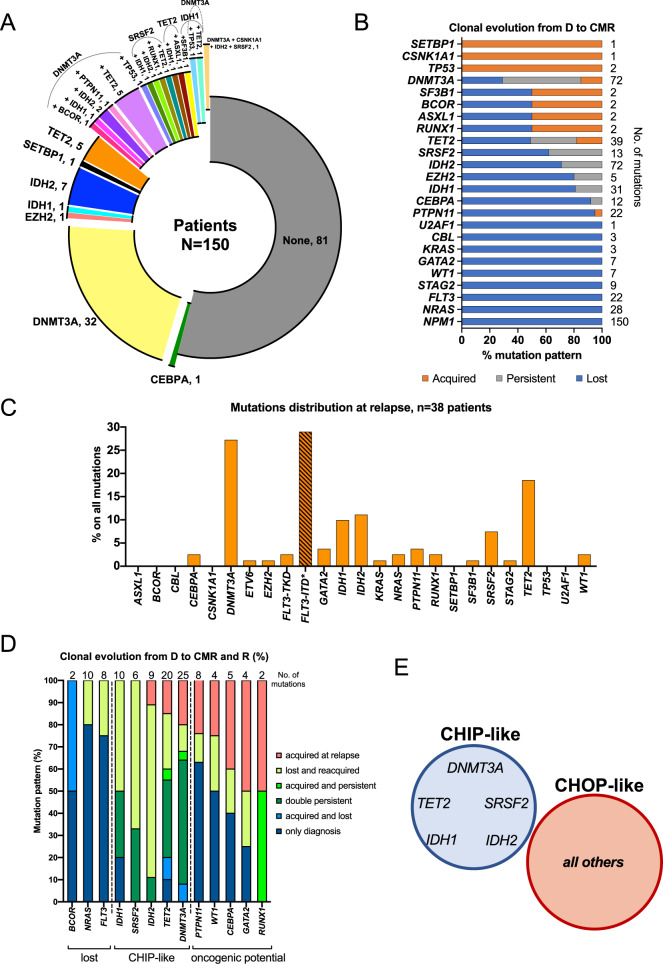
Fig. 2. Clonal evolution defines persistent CHIP and CHOP-like mutations in NPM1mut AML.
A Donut plot depicting the mutational status of patients at complete molecular remission (CMR). 81/150 patients (54%) had no mutation, whereas 69/150 (46%) had persistency/acquisition of single or combined mutations, for a total of 24 different groups (donut slices). B Clonal evolution analysis of NPM1mut AML from diagnosis to CMR (n = 150 patients). Lost mutations are depicted in blue, persistent mutations in gray and acquired mutations in orange. Three main patterns emerged: mutations that were mostly or completely lost at CMR: NRAS, FLT3-TKD, STAG2, WT1, GATA2, and KRAS (all 100%), PTPN11 (95%), CEBPA (92%), IDH1 (81%), EZH2 (80%), IDH2 (71%); mutations that were mostly or exclusively acquired at CMR (TP53, CSNK1A1 and SETBP1, all 100%), and mutations with a more heterogeneous behavior: SRSF2 (mutation lost in 45% persistent in 38% and acquired in 17% of cases), TET2 (mutation lost in 52%, persistent in 31% and acquired in 17% of cases) and DNMT3A (mutation lost in 29%, persistent in 56% and acquired in 15% of cases). C Mutation frequencies of AML associated genes in relapse samples of 38/52 patients with clinical relapse. The percentage of each gene alteration among all the mutations per timepoint is depicted. *FLT3-ITD mutations were detected by gene scan. D Clonal evolution analysis of NPM1mut AML from diagnosis to CMR and relapse (R) (n = 38 patients) allows for higher temporal resolution and identifies three main patterns: mutations which could either persist at CMR and be lost at R or completely absent at both CMR and R (BCOR, NRAS, FLT3-TKD); CHIP-like mutations present at diagnosis, CMR and R (TET2, IDH1, IDH2, DNMT3A, SRSF2); mutations with oncogenic potential: gained at CMR and persistent at R or acquired de novo at relapse (CEBPA, PTPN11, WT1, GATA2, RUNX1). E DNMT3A, TET2, IDH1, IDH2, and SRSF2 often act as foundation mutations onto which other potentially oncogenic (CHOP) hits arise as later events in AML pathogenesis. Venn diagram showing the novel proposed classification of CHIP-like mutations including: DNMT3A, TET2, IDH1, IDH2, and SRSF2, versus mutations with oncogenic potential (CHOP) in the context of NPM1mut AML.