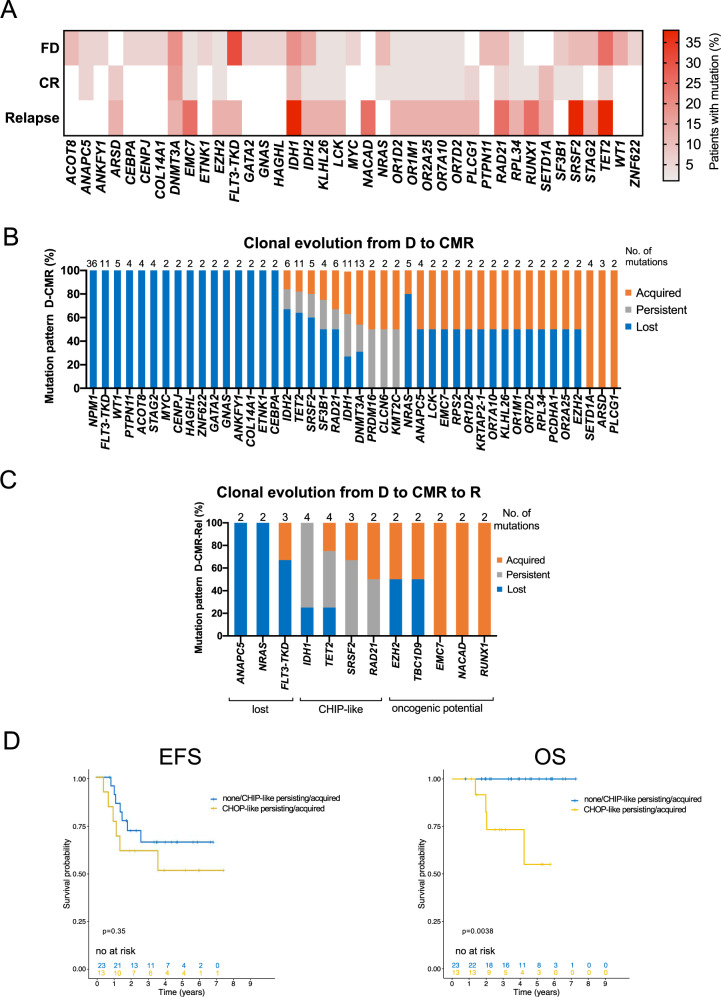
Fig. 5. Mutational analysis of independent WGS cohort recapitulates the panel-seq findings and confirms predictive power of CHOP-like mutations.
A Mutation frequencies of AML associated genes in diagnostic (D), complete molecular remission (CMR) and relapse samples (R). Mutations detected in at least two patients are depicted. B Clonal evolution analysis of NPM1mut AML from diagnosis to CMR (n = 36 patients) of genes mutated in at least two patients. Lost mutations are depicted in blue, persistent mutations in gray and acquired mutations in orange. C Clonal evolution analysis of NPM1mut AML from D to CMR and R (n = 8 patients) allows for higher temporal resolution and identifies three main patterns: mutations which could either persist at CMR and be lost at R or completely absent at both CMR and R (ANAPC5, NRAS, FLT3-TKD); CHIP-like mutations present at D, CMR and R (TET2, IDH1, SRSF2); mutations with oncogenic potential: gained at CMR and persistent at R or acquired de novo at R (RAD21, TBC1D9, EZH2, EMC7, NACAD, RUNX1); D Survival analysis of 36 patients from the WGS cohort stratified by clonal evolution patterns of novel defined CHIP-like mutations (DNMT3A, TET2, ASXL1, SRSF2, IDH1, IDH2): Kaplan–Meier plots depicting event-free survival (EFS, left panel) and overall survival (OS, right panel) of NPM1mut AML patients based on the persistency/acquisition of CHIP vs CHOP like mutations at CMR. P values were calculated with the log-rank test.