Figure 3.
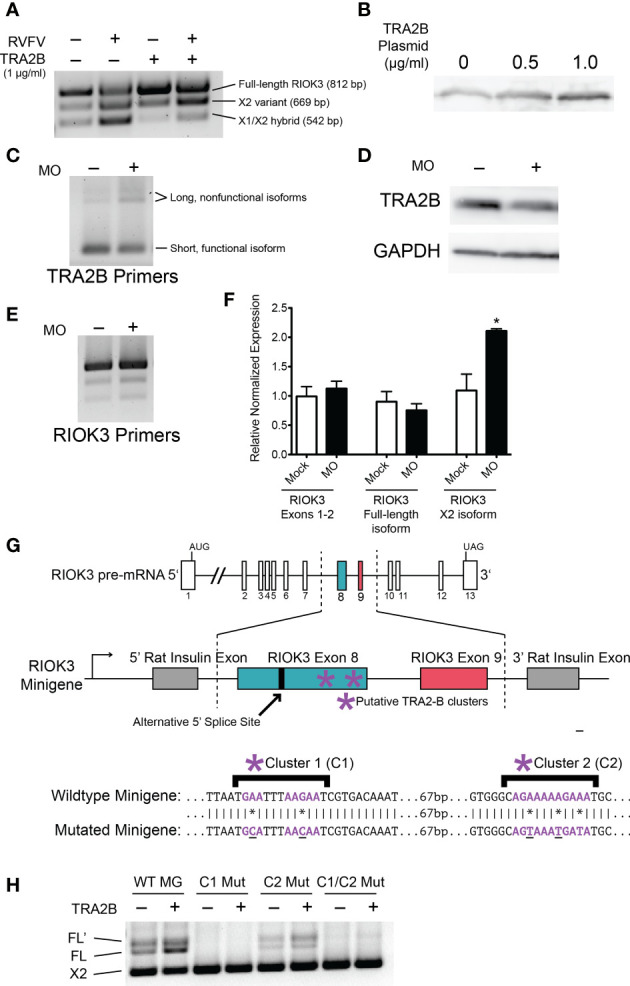
RIOK3 mRNA alternative splicing bias is controlled by TRA2-β. (A) HEK293 cells were transfected with TRA2-β overexpression plasmid or GFP control for 24h, then infected with RVFV MP-12 (MOI = 1) or mock for 24h before harvesting. RT-PCR shows RIOK3 splice isoforms. (B) HEK293 cells were transfected with TRA2-β overexpression plasmid. 24h post-transfection, cells were lysed and a western blot was performed to estimate overexpression. (C) HEK293 cells were treated with TRA2-β poison exon-inducing morpholino oligonucleotides (MO) at 10 μM for 24h, and RT-PCR was performed using TRA2-β primers. (D) HEK293 cells were treated with 5 μM MO for 24h, and lysate was used for a western blot to estimate knockdown of TRA2-β. (E) HEK293 cells were treated with MO as in panel C, and RT-PCR was performed using primers against RIOK3. (F) HEK293 cells were treated with MO as in panel C and RNA was used to perform RT-qPCR. RIOK3 exons 1-2 were used to measure overall expression of the mRNA. Each lane indicates data from biological triplicates and technical duplicates. Asterisk indicates p < 0.05 compared to mock transfection (Student’s t-test). (G) Schematic illustration of RIOK3 splicing minigene. RIOK3 exons 8 and 9 were cloned into pSpliceExpress vector, and primers spanning the splice junction between the rat insulin exon and the RIOK3 exon were used to amplify cDNA for RT-PCR. Lower panel shows mutations in putative TRA2-β binding sites. (E) RT-PCR using splice junction-spanning primers to amplify minigene cDNA.
