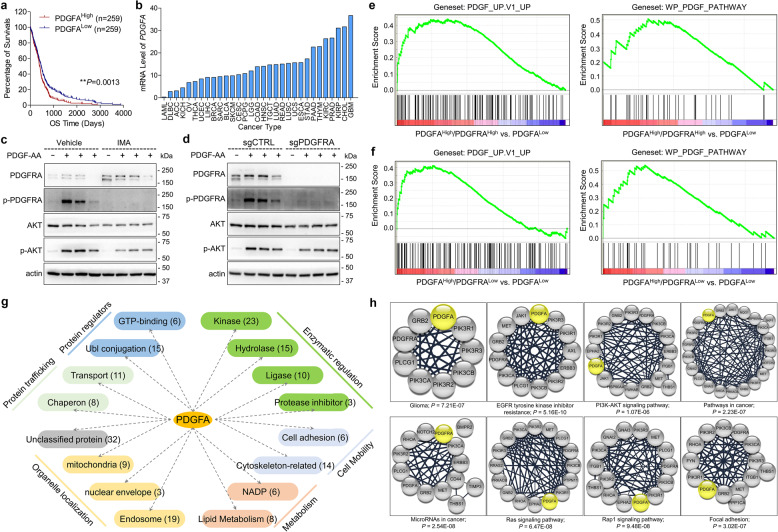
Fig. 1.
PDGFRA is not necessary for PDGFA function in GBM cells. a Kaplan–Meier survival analysis of cases with PDGFAHigh vs. PDGFALow from TCGA_GBM database. b PDGFA gene expression in TCGA PanCancer databases. c PDGFA-induced temporal expression of indicated proteins in LN18 cells pre-treated with vehicle or IMA. β-actin is used as loading control. d PDGFA-induced temporal expression of indicated proteins in LN18 cells transfected with control CRISPR/Cas9 or CRISPR/Cas9 targeting PDGFRA. e, f Enrichment of PDGF signaling-related genesets for TCGA_GBM cases with PDGFAHigh/PDGFRAHigh vs. PDGFALow or PDGFAHigh/PDGFRALow vs. PDGFALow. g Categorization of PDGFA-associated proteins. h Significantly enriched KEGG pathways involving PDGFA through KEGG analysis on PDGFA interactome.