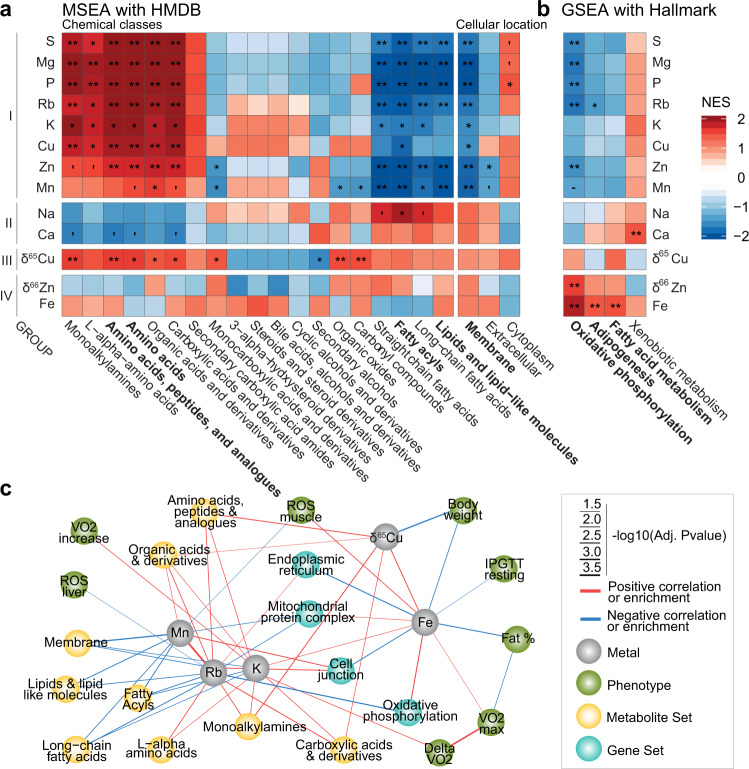
Fig. 4. Enrichment analyses in the liver metabolome and proteome converge towards common associations between ions and mitochondrial and fatty acid metabolism.
a, b Metabolites and proteins were sorted on their correlation with individual metals in the liver, then metabolite or gene -set enrichment analyses (MSEA, (a), GSEA, (b)) were performed. For each metal, the top three gene or metabolite sets are pictured. FDR-corrected p-values.**Adj.P < 0.01, *Adj.P < 0.05, ‘Adj.P < 0.1 (c) Network representation, combining the most significant correlations between metals, between metals and phenotypes (Spearman Rho and limma test, both), as well as the metabolite and gene set enrichment analyses. All tests were two-sided when applicable and p-values are FDR corrected. To limit the number of nodes, only nodes with very significant associations are pictured (Adj.P < 0.01), but all edges with Adj.P < 0.05 are drawn. Source data are provided as a Source Data file.